For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGSMSMNAEQTVAELIARSNRLGSDPKNTNYAGGNTSAKGTDTDPVTGQPVELLWVKGSGGDLGTLTENG LAVLRLDRMRALVDVYPGVEREDEMVAAFDYCLHGRGGAAPSIDTAMHGLVDAAHVDHLHPDSGIALATA ADGEALTKQIFGDRVVWVPWRRPGFQLGLDIADIKRRNPQAIGTILGGHGITAWGSTSAEAEAHSLEIIE TAERYIAENGRPEPFGAPLAGYGALPDDARRAKAAQLAPFIRGLVSTDRPQVGHFTDDPRVLEFLSASEH PRLAALGTSCPDHFLRTKVKPLVLDLPADATVDECKNRLAELHEAYRVDYQAYYDRHATPDSPAIRGADP AIVLIPGVGMFSYGKDKQTARVAGEFYLNAINVMRGAEAISTYAPIDEAEKFRIEYWALEEAKLQRMPKP KPLATRIALVTGAASGIGKAIATRLAAEGACVVIADLDGAKAESAAAEIGNTDVAVGIAADVTDEAAVQA AVDATVLAFGGIDIVVNNAGLSLSKSLLDTTAEDWDLQHNVMARGSFLVSKAAARALIEQKLGGDIIYIS SKNSVFAGPNNIAYSATKADQAHQVRLLAAELGEHGVKVNGINPDGVVRGSGIFAGGWGAKRAAVYGVPE EELGKYYAQRTLLKREVLPEHVANAAFALCTSDFSHTTGLHVPVDAGVAAAFLR
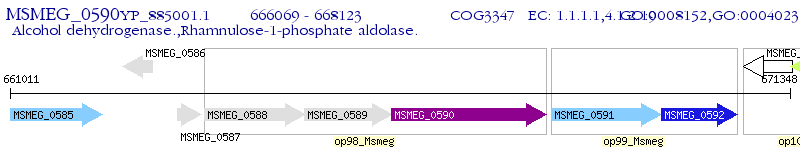
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0590 | rhaD | - | 100% (684) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3607 | - | 3e-30 | 35.18% (253) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4117 | - | 8e-30 | 32.57% (261) | 2-deoxy-D-gluconate 3-dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1976 | - | 2e-31 | 37.88% (264) | short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_2424 | - | 2e-30 | 33.98% (259) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1941 | - | 2e-31 | 37.88% (264) | short-chain type dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_00862 | ephD | 6e-21 | 38.89% (180) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0073c | - | 1e-25 | 34.10% (261) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_2869 | - | 2e-32 | 37.79% (262) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_5088 | fabG | 6e-31 | 37.11% (256) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. thermoresistible (build 8) | TH_3505 | - | 2e-29 | 35.14% (259) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_2885 | - | 3e-32 | 37.79% (262) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_3999 | - | 8e-32 | 45.05% (182) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1976|M.bovis_AF2122/97 --------------------------------------------------
Rv1941|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2869|M.marinum_M --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3999|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0590|M.smegmatis_MC2_155 MGSMSMNAEQTVAELIARSNRLGSDPKNTNYAGGNTSAKGTDTDPVTGQP
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2424|M.gilvum_PYR-GCK --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MAV_5088|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 --------------------------------------------------
Mb1976|M.bovis_AF2122/97 --------------------------------------------------
Rv1941|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2869|M.marinum_M --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3999|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0590|M.smegmatis_MC2_155 VELLWVKGSGGDLGTLTENGLAVLRLDRMRALVDVYPGVEREDEMVAAFD
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2424|M.gilvum_PYR-GCK --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MAV_5088|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 --------------------------------------------------
Mb1976|M.bovis_AF2122/97 --------------------------------------------------
Rv1941|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2869|M.marinum_M --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3999|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0590|M.smegmatis_MC2_155 YCLHGRGGAAPSIDTAMHGLVDAAHVDHLHPDSGIALATAADGEALTKQI
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2424|M.gilvum_PYR-GCK --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MAV_5088|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb1976|M.bovis_AF2122/97 --------------------------------------------------
Rv1941|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2869|M.marinum_M --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3999|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0590|M.smegmatis_MC2_155 FGDRVVWVPWRRPGFQLGLDIADIKRRNPQAIGTILGGHGITAWGSTSAE
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2424|M.gilvum_PYR-GCK --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MAV_5088|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb1976|M.bovis_AF2122/97 --------------------------------------------------
Rv1941|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2869|M.marinum_M --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3999|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0590|M.smegmatis_MC2_155 AEAHSLEIIETAERYIAENGRPEPFGAPLAGYGALPDDARRAKAAQLAPF
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2424|M.gilvum_PYR-GCK --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MAV_5088|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb1976|M.bovis_AF2122/97 --------------------------------------------------
Rv1941|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2869|M.marinum_M --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3999|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0590|M.smegmatis_MC2_155 IRGLVSTDRPQVGHFTDDPRVLEFLSASEHPRLAALGTSCPDHFLRTKVK
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2424|M.gilvum_PYR-GCK --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MAV_5088|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb1976|M.bovis_AF2122/97 --------------------------------------------------
Rv1941|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2869|M.marinum_M --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3999|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0590|M.smegmatis_MC2_155 PLVLDLPADATVDECKNRLAELHEAYRVDYQAYYDRHATPDSPAIRGADP
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2424|M.gilvum_PYR-GCK --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MAV_5088|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb1976|M.bovis_AF2122/97 --------------------------------------------------
Rv1941|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2869|M.marinum_M --------------------------------------------------
MUL_2885|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3999|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0590|M.smegmatis_MC2_155 AIVLIPGVGMFSYGKDKQTARVAGEFYLNAIN---VMRGAEAISTYAPID
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2424|M.gilvum_PYR-GCK --------------------------------------------------
TH_3505|M.thermoresistible__bu --------------------------------------------------
MAV_5088|M.avium_104 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb1976|M.bovis_AF2122/97 --------------------MNHPDLAGKVAIVTGAGAGIGLAVARRLAD
Rv1941|M.tuberculosis_H37Rv --------------------MNHPDLAGKVAIVTGAGAGIGLAVARRLAD
MMAR_2869|M.marinum_M --------------------MRYPDLAGKATIVTGAGAGIGLAIAERLAA
MUL_2885|M.ulcerans_Agy99 --------------------MRYPDLAGKATIVTGAGAGIGLAIAERLAA
Mvan_3999|M.vanbaalenii_PYR-1 --------------------MTYPDLADKAVIVTGAAAGIGLAIAERLAA
MSMEG_0590|M.smegmatis_MC2_155 EAEKFRIEYWALEEAKLQRMPKPKPLATRIALVTGAASGIGKAIATRLAA
MAB_0073c|M.abscessus_ATCC_199 ----------------------MTRLGGRVAVVTGAGKGMGRTHCERLAE
Mflv_2424|M.gilvum_PYR-GCK ----------------MAITASDILLTGRVAVVTGGGAGIGRGIAQGLAA
TH_3505|M.thermoresistible__bu ----------------LTIAPADILLTDRVAVVTGGGAGLGRGIAAGFAA
MAV_5088|M.avium_104 --------------------MAESLLLGKVAVVTGAGQGIGREIARALHR
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
: . ***.. *:* . :
Mb1976|M.bovis_AF2122/97 EGCHVLCADIDG--DAADAAATKIGCG---AAACRVDVSDEQQIIAMVDA
Rv1941|M.tuberculosis_H37Rv EGCHVLCADIDG--DAADAAATKIGCG---AAACRVDVSDEQQIIAMVDA
MMAR_2869|M.marinum_M EGAAVLCADIDG--ASAEQAAAKIGSG---AVGYQVDVSAEEQVAGMVEA
MUL_2885|M.ulcerans_Agy99 EGAAVLCADIDG--ASAEQAAAKIGSG---AVGYQVDVSAEEQVAGMVEA
Mvan_3999|M.vanbaalenii_PYR-1 ERCRVVCADIDG--AAADAAAAAIGNG---AVGYAVDVSDEPQVVGMVEA
MSMEG_0590|M.smegmatis_MC2_155 EGACVVIADLDG--AKAESAAAEIGNTDV-AVGIAADVTDEAAVQAAVDA
MAB_0073c|M.abscessus_ATCC_199 EGSDVIAIDLPGSDDLAQTAAAVEEHGRR-CVWAFADVRDAAALTEAIDT
Mflv_2424|M.gilvum_PYR-GCK FGASVAIWERDP--DTCAAAAAAIG-----GLGLEVDVRDAAAVDTALDR
TH_3505|M.thermoresistible__bu FGARVAIWERDP--DSCARTAEEIG-----ALGIVTDVRDSAAVSAALRQ
MAV_5088|M.avium_104 HGARVVLADLDG--SAARSAAAQIDDSGANCTGLACDVTSEEQVGALVAG
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDE--AAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQ
. * : . :* ** :
Mb1976|M.bovis_AF2122/97 CVAAFGGVDKLVANAGVVHLAS-LIDTTVEDFDRVIAINLRGAWLCTKHA
Rv1941|M.tuberculosis_H37Rv CVAAFGGVDKLVANAGVVHLAS-LIDTTVEDFDRVIAINLRGAWLCTKHA
MMAR_2869|M.marinum_M CVTSFGGLDKLVANAGVVHLAP-ILDTAVEDFDRVIGINLRGAWLCTKHA
MUL_2885|M.ulcerans_Agy99 CVTSFGGLDKLVANAGVVHLAP-ILDTAVEDFDRVIGINLRGAWLCTKHA
Mvan_3999|M.vanbaalenii_PYR-1 CVAAFGGVDKLVANAGVVHFAP-LTETTVDDFDRIIGINLRGAWLCTKHA
MSMEG_0590|M.smegmatis_MC2_155 TVLAFGGIDIVVNNAGLSLSKS-LLDTTAEDWDLQHNVMARGSFLVSKAA
MAB_0073c|M.abscessus_ATCC_199 GVRMLGRLDIVVANAGVYDGLAPSCELTEDSWRRALDVNLTGAWHTVKAG
Mflv_2424|M.gilvum_PYR-GCK TERELGTVSILVNNAGGTFNSP-LLDTSENGWDALYRSNLRHVLLCTQRV
TH_3505|M.thermoresistible__bu TTEQLGSPTILVNNAGGTFSSP-LLETSENGWDALYRANLRHVLLCTQQV
MAV_5088|M.avium_104 TVREHGGLDVFVNNAGITRDAS-LKRMTVADFDAVIAVHLRGTWLGVREA
MLBr_00862|M.leprae_Br4923 VSAKHGVPDIVVNNAGIGQAGR-FLDTPPEEFDRVLAVNLGGVVNCCRAF
* .* *** . : :
Mb1976|M.bovis_AF2122/97 APRMIERG-GGAIVNLSSLAGQVAVGGTGAYGMSKAGIIQLSRITAAELR
Rv1941|M.tuberculosis_H37Rv APRMIERG-GGAIVNLSSLAGQVAVGGTGAYGMSKAGIIQLSRITAAELR
MMAR_2869|M.marinum_M APKMVERG-GGAIVNMSSLAGHIGVGGTAAYGMSKAGISHLSRVTAAELR
MUL_2885|M.ulcerans_Agy99 APKMVERG-GGAIVNMSSLAGHIGVGGTAAYGMSKAGISHLSRVTAAELR
Mvan_3999|M.vanbaalenii_PYR-1 APRMAERG-GGAIVNMSSLGGLVAAAGTAAYGMSKAGIIHLSRITAAEMR
MSMEG_0590|M.smegmatis_MC2_155 ARALIEQKLGGDIIYISSKNSVFAGPNNIAYSATKADQAHQVRLLAAELG
MAB_0073c|M.abscessus_ATCC_199 AAHLLD---GGSVIIISSTNGLKGTANTAHYTASKHAVVGLARTAANELG
Mflv_2424|M.gilvum_PYR-GCK ARRLVAAEQTGSVINVTSIEGSRAAPGYAAYAAAKAGVVNYTKTASFELA
TH_3505|M.thermoresistible__bu GRQLVAAGQPGSIVNITSIEGARAAPGYAAYAAAKAGVINYTRTAAFELA
MAV_5088|M.avium_104 AAVMRERK-TGSIVNMSSLSGKAGNPGQTNYSAAKAGIVGLTKAAAKELA
MLBr_00862|M.leprae_Br4923 GQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELD
. : * :: ::* . . * :* *:
Mb1976|M.bovis_AF2122/97 SSGIRSNTLLPAFVDTPMQQT-------AMAMFDGALGAGGARSMIARLQ
Rv1941|M.tuberculosis_H37Rv SSGIRSNTLLPAFVDTPMQQT-------AMAMFDGALGAGGARSMIARLQ
MMAR_2869|M.marinum_M SANVRSNALLPAFVDTPMQQS-------AMAMFDEALGTGGANTMIARLQ
MUL_2885|M.ulcerans_Agy99 SANVRSNALLPAFVDTPMQQS-------AMAMFDEVLGTGGANTMIARLQ
Mvan_3999|M.vanbaalenii_PYR-1 SANIRSNALLPAFVDTPMQQT-------AMTMFDEALGDGGAATMIGRLQ
MSMEG_0590|M.smegmatis_MC2_155 EHGVKVNGINPDGVVRGSGIFAGGWGAKRAAVYGVPEEELGKYYAQRTLL
MAB_0073c|M.abscessus_ATCC_199 PQGIRVNTVHPGAVSTGMILNPAVIARLRPDLEHPTVEDAAEALAARTLL
Mflv_2424|M.gilvum_PYR-GCK PYGIRVNGLAPDLTMTEG-------------LQKLAFGELSGTVARMVPM
TH_3505|M.thermoresistible__bu PHRIRVNAIAPDIAVTEG-------------LQRMSPDGIPPEVGHAIPL
MAV_5088|M.avium_104 HHNVRVNAIQPGLIRTPM--------------TAAMPPDVFAEREAAVPM
MLBr_00862|M.leprae_Br4923 AVGVGLTTICPGVIDTNIIQSTR---IDTPTAKQERVRDRRGQLDMMFRL
: . : *
Mb1976|M.bovis_AF2122/97 GR-MAAPEEMAGIVVFLLSDDASMITGTTQIADGGTIAALW---------
Rv1941|M.tuberculosis_H37Rv GR-MAAPEEMAGIVVFLLSDDASMITGTTQIADGGTIAALW---------
MMAR_2869|M.marinum_M GR-MARPEEMASIVAFLLSDDSSMITGTTQIADGGTIAALW---------
MUL_2885|M.ulcerans_Agy99 GR-MARPEEMARIVAFLLPDDSSMITGTTQIADGGTIAALW---------
Mvan_3999|M.vanbaalenii_PYR-1 GR-MAGPAEMAGVVAFLLSDAASMVNGTAQLADGGTLAALW---------
MSMEG_0590|M.smegmatis_MC2_155 KR-EVLPEHVANAAFALCTSDFSHTTGLHVPVDAGVAAAFLR--------
MAB_0073c|M.abscessus_ATCC_199 PVPWVEAVDISNAVVFLASDEARYITGTQLVVDAGLTQKA----------
Mflv_2424|M.gilvum_PYR-GCK RR-AGHVDEMASAAVFLASDMASYITGQTIHVDGGTEAAGGWYFHPDNGA
TH_3505|M.thermoresistible__bu GR-PGHVDEIAGAAVFLASEMSSYITGQTLHVDGGTQAAGGWYHHPRTGA
MAV_5088|M.avium_104 KR-AGEPEEVAGAVVFLASELSSYITGTVLGVGGGRYM------------
MLBr_00862|M.leprae_Br4923 RR-YGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTARI
: . . . *
Mb1976|M.bovis_AF2122/97 ---------
Rv1941|M.tuberculosis_H37Rv ---------
MMAR_2869|M.marinum_M ---------
MUL_2885|M.ulcerans_Agy99 ---------
Mvan_3999|M.vanbaalenii_PYR-1 ---------
MSMEG_0590|M.smegmatis_MC2_155 ---------
MAB_0073c|M.abscessus_ATCC_199 ---------
Mflv_2424|M.gilvum_PYR-GCK PTLGPA---
TH_3505|M.thermoresistible__bu PTLGPGPVR
MAV_5088|M.avium_104 ---------
MLBr_00862|M.leprae_Br4923 RVI------