For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAARDRLMAAALKLFAAKGYAATSVAEIQQQAGLAPGSGALYKHFGSKRELLEAAITHRIDNIVAAREQY DAEKPRTVEDAVRSAGQLIWTNLTQSEELLRVMLREPDELGDLDEKTWQVITDNAYQRFADELAASNRAG RTEIPDPEATAAVAIASLSYVATLHALNGRTPGNVDHDRFFEAWVAQTVAAIEQHRTDNQTDNR*
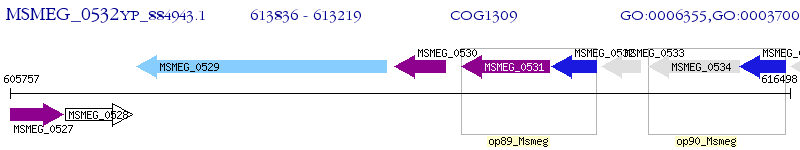
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0532 | - | - | 100% (205) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2193 | - | 1e-08 | 26.50% (200) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1935 | - | 6e-07 | 24.14% (174) | TetR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1561 | - | 4e-09 | 27.71% (166) | transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4672 | - | 1e-07 | 26.84% (190) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1534 | - | 4e-09 | 27.71% (166) | transcriptional regulator |
| M. leprae Br4923 | MLBr_00815 | - | 1e-05 | 34.85% (66) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3313c | - | 1e-14 | 29.44% (197) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0150 | - | 3e-84 | 77.44% (195) | TetR family transcriptional regulator |
| M. avium 104 | MAV_0146 | - | 2e-87 | 75.24% (210) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2855 | - | 3e-08 | 27.44% (164) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_4947 | - | 2e-83 | 76.41% (195) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4909 | - | 4e-10 | 31.79% (151) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0150|M.marinum_M ---------------------------MVSEYSLTMSTSGR-DRLLSAAL
MUL_4947|M.ulcerans_Agy99 -----------------------------------MSTSGR-DRLLSAAL
MAV_0146|M.avium_104 -----------------------------------MLSSGR-DRLLSAAL
MSMEG_0532|M.smegmatis_MC2_155 ------------------------------------MSAAR-DRLMAAAL
Mb1561|M.bovis_AF2122/97 ---MSRASARRRRAVSD-----------------EDKSQRR-DEILAAAK
Rv1534|M.tuberculosis_H37Rv ---MSRASARRRRAVSD-----------------EDKSQRR-DEILAAAK
Mflv_4672|M.gilvum_PYR-GCK MSELAKTAPRRGGQPAN-GTGSSGAAR---RGNRLPRDERR-GQLLAAAS
MLBr_00815|M.leprae_Br4923 MNNLAKAAQRRTVRPIDRGRLSDDIATPNQRSNRLSRDKRR-GQLLIVAS
MAB_3313c|M.abscessus_ATCC_199 ----------------------------------MGEISTK-ERLVTEAM
TH_2855|M.thermoresistible__bu LSQSPDNATSSSRAGSGAGSGAGSGAEPIGRDTPRRRPRIRPEEIVSAAA
Mvan_4909|M.vanbaalenii_PYR-1 MNEAHRDAHAASTAPVS----------------RLQRRKMRTRMALIHAA
: : *
MMAR_0150|M.marinum_M KLFAAKGYAATSVADIQRCSGLAPGSGALYKHFGSKHELLEAAVEHRIDS
MUL_4947|M.ulcerans_Agy99 KLFAVKGYAATSVADIQRCSGLAPGSGALYKHFGSKHELLEAAVEHRIDS
MAV_0146|M.avium_104 RLFAAKGYAATSVADIQRESGLAPGSGALYKHFGSKRELLEAAVAHRIES
MSMEG_0532|M.smegmatis_MC2_155 KLFAAKGYAATSVAEIQQQAGLAPGSGALYKHFGSKRELLEAAITHRIDN
Mb1561|M.bovis_AF2122/97 IVFAHKGFHATTVADIAKQAGLAYG--LIYWYFDSKDDLFHALMAGEEEA
Rv1534|M.tuberculosis_H37Rv IVFAHKGFHATTVADIAKQAGLAYG--LIYWYFDSKDDLFHALMAGEEEA
Mflv_4672|M.gilvum_PYR-GCK EVFVDRGYHAAGMDEIADRAGVSKP--VLYQHFSSKVELYLAVLQRHVDN
MLBr_00815|M.leprae_Br4923 DVFVDHGYHAAGMDEIADRAGVSKP--VLYQHFSSKLELYLAVLERHMNN
MAB_3313c|M.abscessus_ATCC_199 RLFGEQGYKATSVAQIEKAAGLTPGSGGLYHHFKSKEALLEAGIDRQLDR
TH_2855|M.thermoresistible__bu EFFAREGYANVGMRDVAEAVGIRGAS--LYHHFGSKEEILYAICLTVTKE
Mvan_4909|M.vanbaalenii_PYR-1 QGFIAEGKLAVPVLEITQAADVGMG--SFYNHFDSKDELFEAAVTEALDG
* .* . : :: .: :* :* ** : * .
MMAR_0150|M.marinum_M IVAAREQYDAG----EPGGVEQAVRTAGHLIWTNLKQSEDLLKVMLREPG
MUL_4947|M.ulcerans_Agy99 IVAAREQYDAG----EPGGVEQAVRTAGHLIWTNLKQSEDLLKVMLREPG
MAV_0146|M.avium_104 IVTAREQYDAG----QPGSVEQAVRTAGQLIWNNLKQSEDLLKVMLREPD
MSMEG_0532|M.smegmatis_MC2_155 IVAAREQYDAE----KPRTVEDAVRSAGQLIWTNLTQSEELLRVMLREPD
Mb1561|M.bovis_AF2122/97 LRAHVAAELARVGGSTEAPLRALLQAAVQATFEFFETDKATVKLLFRDAY
Rv1534|M.tuberculosis_H37Rv LRAHVAAELARVGGSTEAPLRALLQAAVQATFEFFETDKATVKLLFRDAY
Mflv_4672|M.gilvum_PYR-GCK LVSGVRQALR-----TTTDNRQRVRAAVQAFFDFIEHDSQGYRLIFENDY
MLBr_00815|M.leprae_Br4923 LVSSVQQALR-----TTTNNRQRLHAAVQAFFDFIEHDGQGYRLIFENDN
MAB_3313c|M.abscessus_ATCC_199 RRAMRDIRALFG---GLGDLRNELTMMGRYVLAVLDEESQLLQIASSVPS
TH_2855|M.thermoresistible__bu PNELNLPLLD-----AAGTPAERLSALIRAHLEHLIRRRVEHLVALHDIN
Mvan_4909|M.vanbaalenii_PYR-1 FGALLDSYTA-----AVADPAEAFATSFRLSGRLFRIRPQEAKLLLAN--
. : : :
MMAR_0150|M.marinum_M ALGDLDEKTWQIITDNAYQRFADELAASNRSGRTRIPDPEAAASVAIASL
MUL_4947|M.ulcerans_Agy99 ALGDLDEKTWQIITDNAYQRFADELAASNRSGRTRIPDPEAAASVAIASL
MAV_0146|M.avium_104 ELGDLDEKTWQVITDNAYQRFADELAASNRAGRTSIPDPEAAAAVAIGSL
MSMEG_0532|M.smegmatis_MC2_155 ELGDLDEKTWQVITDNAYQRFADELAASNRAGRTEIPDPEATAAVAIASL
Mb1561|M.bovis_AF2122/97 ALGGRFEEHLGGIYERFIDDIEAVVVAAQRRG-EVVEAPSRMAAYTLAAL
Rv1534|M.tuberculosis_H37Rv ALGGRFEEHLGGIYERFIDDIEAVVVAAQRRG-EVVEAPSRMAAYTLAAL
Mflv_4672|M.gilvum_PYR-GCK VTEPQVAAQVKVATESCTDAVFDLISHDSG----LEPHRARMIAVGLVSI
MLBr_00815|M.leprae_Br4923 ITEPRVAAQVRVATESCIDAVFALISADSR----LDPHRARMVAVGLVGI
MAB_3313c|M.abscessus_ATCC_199 GRPARLDHAYAALFDGLYGELSDWVQGWAP--DISKQDAESIGVLGVNAL
TH_2855|M.thermoresistible__bu SLTPEHRAIIDDLRRYYQRRVRDVIVAGVRSGEFDVPDPKLAALLILDAL
Mvan_4909|M.vanbaalenii_PYR-1 ------GSSLILSEHGLSPRALRDITAAIDKGRFKITDPELGLAIAAGAL
: .:
MMAR_0150|M.marinum_M SYAATLQALTGRSPGNIDDSRYFEAWVNQT--------------VSMLSQ
MUL_4947|M.ulcerans_Agy99 SYALTLQALTGRSPGNIDDSRYFEAWVNQT--------------VSMLSQ
MAV_0146|M.avium_104 SYAATLQALTGRLPGNTDEDRYFEAWVNQT--------------VSVLAQ
MSMEG_0532|M.smegmatis_MC2_155 SYVATLHALNGRTPGNVDHDRFFEAWVAQT--------------VAAIEQ
Mb1561|M.bovis_AF2122/97 VGQLAHRRLNTDDN--VTAAQVADFVVSLV--------------LDGLRP
Rv1534|M.tuberculosis_H37Rv VGQLAHRRLNTDDN--VTAAQVADFVVSLV--------------LDGLRP
Mflv_4672|M.gilvum_PYR-GCK SVDSARYWLNNERP--IDKDDAVEGTVQFI--------------WGGLSH
MLBr_00815|M.leprae_Br4923 SVDCARYWLDTNRP--ISKSDAVEGTVQFA--------------WGGLSH
MAB_3313c|M.abscessus_ATCC_199 LGKRATSAVFRGAARDIADEDYVVEWTALL--------------AMRIES
TH_2855|M.thermoresistible__bu NGVSGWFHPDRGHS----LPDVIAAYTEMI--------------VERVLR
Mvan_4909|M.vanbaalenii_PYR-1 YGLGTLLTQRPDRDGASATDEVTASVLRMFGMTSQQARGLCRHPLPDLSS
:
MMAR_0150|M.marinum_M YTN-PAPSRKP--------
MUL_4947|M.ulcerans_Agy99 YTN-PAPSRKPQADNKFQE
MAV_0146|M.avium_104 YTNRRKPLNDNDNDSGAQT
MSMEG_0532|M.smegmatis_MC2_155 HRTDNQTDNR---------
Mb1561|M.bovis_AF2122/97 RALAVGARGGRAART----
Rv1534|M.tuberculosis_H37Rv RALAVGARGGRAART----
Mflv_4672|M.gilvum_PYR-GCK VPLTRS-------------
MLBr_00815|M.leprae_Br4923 VPLTRL-------------
MAB_3313c|M.abscessus_ATCC_199 LR-----------------
TH_2855|M.thermoresistible__bu ARIS---------------
Mvan_4909|M.vanbaalenii_PYR-1 LQDTRAARPTPIPPAGVT-