For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNVEINQRYDFIVCGSGSSGSVVARRLAENPDADVLLLEAGGSDDVPEVQQAVQWPENLGSPRDWQFEAQ ANPHLNGRAIPMNMGKVLGGGSSINVMMWSRGHRNDWDFFAAEADDEGWGYEAVLDIYRRIEDWQGKPDP QYRGTDGNVYVQPAPDPNPIADAAVAAAASVGIPAFDHPNGAMMEGAGGAALSDIRVRDGIRQSVFRSYV HPYLGRPNLTVLTGAVVTRVTLDGHRASGVEFVRDGVTFRVAAGSEVVLGLGAINTPKVLMLSGIGDSAE LARHGIKTVVHLPGVGRNLQDHPAFGCVWEYDVALEPRNTGSEATYFAKSDSRLDTPDLQTCQIEAPFAS AETIARFGLPQAAWTMFAGIVRPESRGSVRLTGCSPDDPVRIDANMLSDPADMTAAMIAVELCREIGNAA PLSAHAKREVMPGNLKGRDLERFIRDAAVSYWHQSGTAKMGRDEMSVVDGNLRVHGIERLRIADASIMPR ITTGNTMAPCVVIGERAAQLIRDTYAAQEPAHE
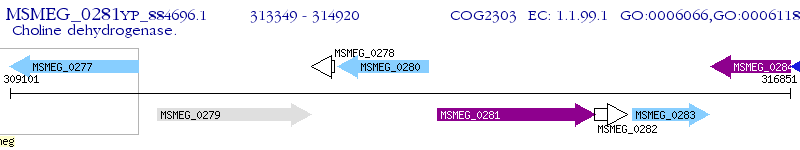
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0281 | - | - | 100% (523) | choline dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3432 | - | e-144 | 55.73% (445) | choline dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3444 | - | 5e-78 | 37.20% (508) | choline dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1310 | - | 3e-79 | 38.27% (533) | dehydrogenase FAD flavoprotein GMC oxidoreductase |
| M. gilvum PYR-GCK | Mflv_3230 | - | 2e-70 | 33.51% (567) | glucose-methanol-choline oxidoreductase |
| M. tuberculosis H37Rv | Rv1279 | - | 4e-79 | 38.27% (533) | dehydrogenase FAD flavoprotein GMC oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0250 | - | 8e-69 | 35.42% (528) | GMC-type oxidoreductase |
| M. marinum M | MMAR_4427 | - | 0.0 | 64.57% (508) | dehydrogenase fad flavoprotein GMC oxidoreductase |
| M. avium 104 | MAV_4980 | - | 2e-65 | 35.29% (442) | choline dehydrogenase |
| M. thermoresistible (build 8) | TH_3275 | - | 4e-75 | 36.05% (552) | PUTATIVE putative oxidoreductase, GMC family protein |
| M. ulcerans Agy99 | MUL_4004 | - | 2e-78 | 37.59% (532) | dehydrogenase fad flavoprotein GMC oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_1606 | - | 0.0 | 72.80% (511) | glucose-methanol-choline oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1310|M.bovis_AF2122/97 ---------------MDT--------QSDYVVVGTGSAGAVVASRLSTDP
Rv1279|M.tuberculosis_H37Rv ---------------MDT--------QSDYVVVGTGSAGAVVASRLSTDP
MUL_4004|M.ulcerans_Agy99 ---------------MDI--------ECDYVVVGTGSAGAAVVNRLSADS
MAB_0250|M.abscessus_ATCC_1997 -------------MVEDL--------AADFVIVGAGSAGAPLATRLSERT
Mflv_3230|M.gilvum_PYR-GCK ----------------MP--------EYDYIIVGAGSAGCLLANRLSADP
TH_3275|M.thermoresistible__bu -------MGVVMMGRRSN--------LHDYIVVGAGSSGCTVAGRLAEGK
MAV_4980|M.avium_104 --------------------------------------------------
MSMEG_0281|M.smegmatis_MC2_155 ----------MNVEINQR---------YDFIVCGSGSSGSVVARRLAENP
Mvan_1606|M.vanbaalenii_PYR-1 MGDHGHTDLAERIRLNQNRLRQNLRPQYDFIVCGAGSAGSVIARRLAENP
MMAR_4427|M.marinum_M ---------MCSDPIAAR---------YDFVVCGSGSAGSVVARRLSENS
Mb1310|M.bovis_AF2122/97 ATTVVALEAGPRDKNRFIGVPAAFS-KLFRSEIDWDYLTEPQPELDGREI
Rv1279|M.tuberculosis_H37Rv ATTVVALEAGPRDKNRFIGVPAAFS-KLFRSEIDWDYLTEPQPELDGREI
MUL_4004|M.ulcerans_Agy99 GTSVVALEAGPSDKNRFIGIPAAFP-KLFRSEVDWDYTTEPEPQLGGREI
MAB_0250|M.abscessus_ATCC_1997 NDQVLVLEAGPKDKDMGIHIPAAFS-KLFRSDVDWDYLTEPQPQLNNRQI
Mflv_3230|M.gilvum_PYR-GCK DHRVLLIEAGGKDDWFWIKVPVGYLYTIANPRTDWCYTTEPDPGLAGRSI
TH_3275|M.thermoresistible__bu RHSVLVLEAGGSDLSPFIRIPAGLM--RVNPRNYWHFRLEPDPTRNGAVD
MAV_4980|M.avium_104 -----------------------------------------MQTAIGRAV
MSMEG_0281|M.smegmatis_MC2_155 DADVLLLEAGGSDDVPEVQQAVQWPENLGSPR-DWQFEAQANPHLNGRAI
Mvan_1606|M.vanbaalenii_PYR-1 DVSVLLLEAGGDDDDESVVRADQWPLNLGTSR-DWNFVAEASPHVNGRSL
MMAR_4427|M.marinum_M NVAVLLLEAGGTDDVAEVMDPAQWPVNLGSAR-DWGFASAPSAHLNGRSI
.
Mb1310|M.bovis_AF2122/97 YWPRGKVLGGSSSMNAMMWVRGFASDYDEWAARAG-PRWSYA------DV
Rv1279|M.tuberculosis_H37Rv YWPRGKVLGGSSSMNAMMWVRGFASDYDEWAARAG-PRWSYA------DV
MUL_4004|M.ulcerans_Agy99 YWPRDKVLGGSSSMNAMMWVRGFAADYDEWARLAG-SHWSYG------EL
MAB_0250|M.abscessus_ATCC_1997 YWPRGKTLGGSSSMNAMMWVRGFAADYDDWAQVAG-EQWSFA------NI
Mflv_3230|M.gilvum_PYR-GCK LYARGRVVGGSSSINAMIHMRGQASDYELWAQATGDERWLWGGPDRPGDT
TH_3275|M.thermoresistible__bu TWAAGRVVGGGSSVNAMLWVRGNRADFDTWAKLGA-QGWDYE------SL
MAV_4980|M.avium_104 PFARGRGIGGSSTINAMVFARGHRDSYSRWRDNGA-AGWSFD------EL
MSMEG_0281|M.smegmatis_MC2_155 PMNMGKVLGGGSSINVMMWSRGHRNDWDFFAAEADDEGWGYE------AV
Mvan_1606|M.vanbaalenii_PYR-1 PMNMGKVLGGGSSINVMMWSRGHRSDWDFFAAEASDPGWGYD------AV
MMAR_4427|M.marinum_M PLSMGKVLGGGSSINVMAWARGHRNDWAHFAAEAGDPSWGYE------SV
.: :**.*::*.* ** .: : * :
Mb1310|M.bovis_AF2122/97 LGYFRRIENVTAAWHFVSGDDSGVTGPLHISRQRSPRSVTAAWLAAAREC
Rv1279|M.tuberculosis_H37Rv LGYFRRIENVTAAWHFVSGDDSGVTGPLHISRQRSPRSVTAAWLAAAREC
MUL_4004|M.ulcerans_Agy99 LGYFRQIEDVTAAWHFVNGDDSGVTGPLQISRQRSPSPLTAAWLAATRDC
MAB_0250|M.abscessus_ATCC_1997 APYFKRIEAVEG----ATESDEGTDGALKISKQRSPRSSTAAWLEAVKEA
Mflv_3230|M.gilvum_PYR-GCK LAIYKDLEDYFG----GADDWHGARGEIRVERPRVRWKILDAWQAAAAES
TH_3275|M.thermoresistible__bu LPAMRSLETWEG----GADRYRGGTGPQHVSRIRLHHPLVQKWLTGAAQT
MAV_4980|M.avium_104 LPYFKRSESTCG----KDPSLRGQDGPMAVAPAAHPNPVLVACLCAAIQQ
MSMEG_0281|M.smegmatis_MC2_155 LDIYRRIEDWQGK---PDPQYRGTDGNVYVQPAPDPNPIADAAVAAAASV
Mvan_1606|M.vanbaalenii_PYR-1 LDIYRRIEDWHGS---PDPDYRGSGGEVFVQPAPDPNPVAPAAVDAAAAV
MMAR_4427|M.marinum_M LDVYRRIEDWHGR---CDGDRRGVGGPVFVQTAPDPNPIATAVVDGAGAI
: * . * * : ..
Mb1310|M.bovis_AF2122/97 GFAAARP-NSPRPEG---FCETVVTQRRGARFSTADAYLKPAMRRKNLRV
Rv1279|M.tuberculosis_H37Rv GFAAARP-NSPRPEG---FCETVVTQRRGARFSTADAYLKPAMRRKNLRV
MUL_4004|M.ulcerans_Agy99 GFAPVQP-NSAEPEG---FCETIVTQRRGSRWSTVDAYLRPAMGRRNLSI
MAB_0250|M.abscessus_ATCC_1997 GFDVERA-NTPEPKG---FSETMVCQHGGRRWSTADGYLKPGLRRRNLNV
Mflv_3230|M.gilvum_PYR-GCK GIAPIDEFNRGDNSG---SAYFHVNQRRGRRWSMADAFLHPVAHRPNLTV
TH_3275|M.thermoresistible__bu GMPILADYNGESQFG---AAWSQLSQKNGLRHSAAQAFLAPARRRGEVDV
MAV_4980|M.avium_104 GYPRAADISGGLEVG---FGFTDLNIVDGKRQSAADAYLLPALDRPNLEF
MSMEG_0281|M.smegmatis_MC2_155 GIPAFDHPNGAMMEGAGGAALSDIRVRDGIRQSVFRSYVHPYLGRPNLTV
Mvan_1606|M.vanbaalenii_PYR-1 GIPTFDHPNGKMMEGCGGAAISDMRVRDGLRESVFRSYTYPYLDRPNLTV
MMAR_4427|M.marinum_M GIPVYDSNNGIMMEGPGGASLLDLQLRNRLRESVFRSYVHPYLQQRNLTV
* . * : * * .: * : :: .
Mb1310|M.bovis_AF2122/97 LTGATATRVVIDG---------------DRAVGVEYQSDGQTRIVYARRE
Rv1279|M.tuberculosis_H37Rv LTGATATRVVIDG---------------DRAVGVEYQSDGQTRIVYARRE
MUL_4004|M.ulcerans_Agy99 LTGATATRIVIHG---------------TRATGVEYQREGQHCTVHARRE
MAB_0250|M.abscessus_ATCC_1997 VTDAQARRVLFDG---------------IRAVGVEYVRDGITHTVHARRE
Mflv_3230|M.gilvum_PYR-GCK YTQTQALRLLTDDQVRDDQRCGAWTTATHRATGLRVLKDGQIVDVRARRE
TH_3275|M.thermoresistible__bu ESHAVVDKVIIEN---------------GRAVGVEYVRRGTRKVAYAAKE
MAV_4980|M.avium_104 HSGATVHRLRIRG---------------SRCTGVEYTTAEGRQVTAAARE
MSMEG_0281|M.smegmatis_MC2_155 LTGAVVTRVTLDG---------------HRASGVEFVRDGVTFRVAAGSE
Mvan_1606|M.vanbaalenii_PYR-1 LTGAVVTRVPVHM---------------QRATGVEFEMAGTTHRIDAGVE
MMAR_4427|M.marinum_M LTGATVRRLTFSG---------------SRVTGVEFSHANAIRTVSATQE
: : . :: * *:. * *
Mb1310|M.bovis_AF2122/97 VVLCAGAVNSPQLLMLSGIGDRDHLAEHDIDTVYHAPEVGCNLLDHLVTV
Rv1279|M.tuberculosis_H37Rv VVLCAGAVNSPQLLMLSGIGDRDHLAEHDIDTVYHAPEVGCNLLDHLVTV
MUL_4004|M.ulcerans_Agy99 VVLCGGAVNSPQLLMLSGIGDRSHLADHGIETVYHAPEVGQNLLDHLITP
MAB_0250|M.abscessus_ATCC_1997 VILSGGAINSPQLLMLSGIGEATRLGELGIPVVHDAPQVGQNLLDHLCCP
Mflv_3230|M.gilvum_PYR-GCK VILSAGAIGSPHLMQVSGLGPAELLARHDVPVTVDLPGVGENLQDHLQLR
TH_3275|M.thermoresistible__bu VILSAGALCSPLILLRSGIGPAEELRDHGIDVIADLPGVGRNLQEHPAIT
MAV_4980|M.avium_104 IILCAGAIGSPHLLLASGIGPPNHLRSFRVDVALELPGVGANLQDHPIVP
MSMEG_0281|M.smegmatis_MC2_155 VVLGLGAINTPKVLMLSGIGDSAELARHGIKTVVHLPGVGRNLQDHPAFG
Mvan_1606|M.vanbaalenii_PYR-1 TVLCLGAIHTPKVLMHSGIGDAAELHRLGIAPVQHLPGVGANLQDHPGFG
MMAR_4427|M.marinum_M VVVSLGAVNTPKVLMQSGIGDANDLRGFGIPVVTHLRGVGHNYQDHPRID
:: **: :* :: **:* * : . ** * :*
Mb1310|M.bovis_AF2122/97 LGFDVE----KDSLFA--AEKPGQLISYLLRRRGMLTSNVGEAYGFVRSR
Rv1279|M.tuberculosis_H37Rv LGFDVE----KDSLFA--AEKPGQLISYLLRRRGMLTSNVGEAYGFVRSR
MUL_4004|M.ulcerans_Agy99 LGFDVA----GDSLAA--AEKPMQLINYLLRRRGMLTSNVGEAYGFVRSR
MAB_0250|M.abscessus_ATCC_1997 VGYAVK----SDSLFG--AEKPLQLANYFLRHRGMLTSNVGEAYGFLRSH
Mflv_3230|M.gilvum_PYR-GCK TVYRVRGARTVNTLYRNWITRAGMGIQYLLLRSGPLTMPPSTLGAFAKSD
TH_3275|M.thermoresistible__bu MTFTVTDRTLNQELTP--LSILKGAFDFVLRRRGPATAPLSHAMLFGPPL
MAV_4980|M.avium_104 LVYRAA------------------------RPVPAGRYNHGELLGLIRRH
MSMEG_0281|M.smegmatis_MC2_155 CVWEYD-------------------------VALEPRNTGSEATYFAKSD
Mvan_1606|M.vanbaalenii_PYR-1 CVWEYR-------------------------DPLPPRNTASEATYFWKSD
MMAR_4427|M.marinum_M CVWESH-------------------------EALAPRNNGAEVTVFWKSH
: . :
Mb1310|M.bovis_AF2122/97 PELKLPDLELI-FAPAPFYDEA---------------LVP-PAGHGVVFG
Rv1279|M.tuberculosis_H37Rv PELKLPDLELI-FAPAPFYDEA---------------LVP-PAGHGVVFG
MUL_4004|M.ulcerans_Agy99 PGLDLPDLELI-FAPAPFYDEG---------------LPAHPPGHGVVFG
MAB_0250|M.abscessus_ATCC_1997 PNLTLPDLELI-FAPAPFFDEG---------------IGV-ATEHAIVMG
Mflv_3230|M.gilvum_PYR-GCK PVLASPDLEWH-VQPLSLTKFG---------------EPL-HPYGAITPS
TH_3275|M.thermoresistible__bu EPEGWPQYHVL-MAPLGTEQEKPHATQEEAEQNRYDKQNVSLSRNSIVSA
MAV_4980|M.avium_104 ATGGPPEIQIFGVDSADVPGLG--------------------GADGYVLG
MSMEG_0281|M.smegmatis_MC2_155 SRLDTPDLQTCQIEAPFASAET--------------IARFGLPQAAWTMF
Mvan_1606|M.vanbaalenii_PYR-1 PQLPTPDLQTCQVEVPLTSAEN--------------AARFGVPQAGWTMF
MMAR_4427|M.marinum_M PKLATPDLQICVAEFPLSSPEN--------------AARYGLPAHGWTMC
*: . . .
Mb1310|M.bovis_AF2122/97 PIL-VAPQSRGQITLRSADPHAKPVIEPRYLSDLGGVDRAAMMAGLRICA
Rv1279|M.tuberculosis_H37Rv PIL-VAPQSRGQITLRSADPHAKPVIEPRYLSDLGGVDRAAMMAGLRICA
MUL_4004|M.ulcerans_Agy99 PIL-VAPQSRGQITLRSADPHAKPIIEARYLSDPGGVDRAAMMEGLRICA
MAB_0250|M.abscessus_ATCC_1997 PIL-LKPESSGEITLTSPDPLAKPRIDPRYLSDPAGRDRAAMMFGLRTTA
Mflv_3230|M.gilvum_PYR-GCK VCN-LRPTSRGHVRLADADPLTHPKIFCNYLS--TDADRDIAVRGLRMTR
TH_3275|M.thermoresistible__bu FMSPLHPRGRGTVRLGSPDVDATPVIEHPMLAD--ERDVVDLTAAGRKVR
MAV_4980|M.avium_104 VSV-MQPVSRGRVGLSGPTIQEPPAIDPNYLSD--DRDMGLMIEGLRIGR
MSMEG_0281|M.smegmatis_MC2_155 AGI-VRPESRGSVRLTGCSPDDPVRIDANMLSD--PADMTAAMIAVELCR
Mvan_1606|M.vanbaalenii_PYR-1 AGV-VRPASRGRVWLSSAQPSDPVHIDTGMLSD--PADVAAAVAAVELCR
MMAR_4427|M.marinum_M VGV-VRPQSRGRVRLTGAHPGAPVLIEAEMLAH--PDDMKAAAAGVQLCR
: * . * : * . * *: * . .
Mb1310|M.bovis_AF2122/97 RIAQARPLRDLLGSIARPRNSTELDEATLELALATCSHTLYHPMGTCRMG
Rv1279|M.tuberculosis_H37Rv RIAQARPLRDLLGSIARPRNSTELDEATLELALATCSHTLYHPMGTCRMG
MUL_4004|M.ulcerans_Agy99 RIAQAPALRGLLGPVARPHNSSELDEATLELALTTCSHTLYHPLGTCRMG
MAB_0250|M.abscessus_ATCC_1997 RIAETPSMRAVLGKILRPRNPSDDLEETLVAALENNSHTLYHPVGTCRMG
Mflv_3230|M.gilvum_PYR-GCK KIMASPVLARYRPEEMLP-GPHLVSDDDLQAAARELGTTIFHPVGTCAMG
TH_3275|M.thermoresistible__bu EIFQSPAMRPHVIAENAP-SRGVETDDEWAEFIRNHCFGGSHWVGTARMG
MAV_4980|M.avium_104 KIGMSQALDPWRAEELAP-GDDAVDDAALRSYIQRSVASYFHPVGTCAMG
MSMEG_0281|M.smegmatis_MC2_155 EIGNAAPLSAHAKREVMP---GNLKGRDLERFIRDAAVSYWHQSGTAKMG
Mvan_1606|M.vanbaalenii_PYR-1 EIGNAAPLRPFTKREVMP---GNLRGPALEEFVRNAASTYWHPTGTAKMG
MMAR_4427|M.marinum_M EIGNSAPLRPFAKREVMP---GGLAGAELERFVRDGASTYWHQSGTAKMG
.* : : * * **. **
Mb1310|M.bovis_AF2122/97 SDEAS--------VVDPQLRVRGVDGLRVADASVMPSTVRGHTHAPSVLI
Rv1279|M.tuberculosis_H37Rv SDEAS--------VVDPQLRVRGVDGLRVADASVMPSTVRGHTHAPSVLI
MUL_4004|M.ulcerans_Agy99 SDEAS--------VVDRQLRVRGVDGLRVADASVMPSTVRGHTHAPSVLI
MAB_0250|M.abscessus_ATCC_1997 ADDAS--------VVTPDLTVRGVQGLRVVDASVIPSLIRGHTHAPSVLL
Mflv_3230|M.gilvum_PYR-GCK AFDARGLPRSATTVLDTDCRVFRVAGLRVVDASAMPTITSGNTNAPVMLI
TH_3275|M.thermoresistible__bu ADSDP------DAVVDPQLRVRGVSGLRVADASVMPTLTSGNTNAPCVLI
MAV_4980|M.avium_104 TGPES--------VVDNQLRLHGIDNLRVIDASVMPSLPSNNTAATVYAI
MSMEG_0281|M.smegmatis_MC2_155 RDEMS--------VVDGNLRVHGIERLRIADASIMPRITTGNTMAPCVVI
Mvan_1606|M.vanbaalenii_PYR-1 RDDMS--------VVDGALRVYGVAGLRVADGSIMPRVSTGNTMAPCVVI
MMAR_4427|M.marinum_M RDDMS--------VVDAKLKVHGMENLRVADSSIMPRITTGNTMAPCVVI
*: : : **: *.* :* .:* *. :
Mb1310|M.bovis_AF2122/97 GEKAADLIRS----------
Rv1279|M.tuberculosis_H37Rv GEKAADLIRS----------
MUL_4004|M.ulcerans_Agy99 GAKAADLIRN----------
MAB_0250|M.abscessus_ATCC_1997 GEKAADLILSPQP-------
Mflv_3230|M.gilvum_PYR-GCK AERAARAILH----------
TH_3275|M.thermoresistible__bu GEKAAELIRGR---------
MAV_4980|M.avium_104 AERGADLVRLG---------
MSMEG_0281|M.smegmatis_MC2_155 GERAAQLIRDTYAAQEPAHE
Mvan_1606|M.vanbaalenii_PYR-1 GERAAALLGATHGLRAA---
MMAR_4427|M.marinum_M GERAAEIVKADHGL------
. :.* :