For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDLTDPESLRLLADSLKTAVTQNPSPSGAGLDAALQALGWLDMLDEIPGTAVPLVFAMLGENGVHAPLVN DVVARAAGCPGGGTVPLPFAGGSWVIWSRGDQAGSVLDGELPILRVEPADPLPPAALAAGRSAVGWWLAG ASRAMLELARRHALDRNQFGHPIAAFQAVRHRLAEALVAIEGAEATLRAATDQPDELACLLAKAAAGQAA LTTARHCQQVLAGMGFTAEHQLHHHVKRSLVLDGLLGSSRELTREAGAVLRTKGFAPRLAQL
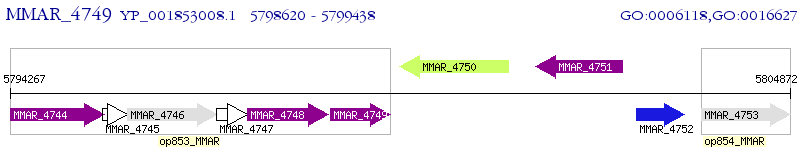
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4749 | - | - | 100% (272) | acyl-CoA dehydrogenase |
| M. marinum M | MMAR_3979 | - | 8e-17 | 39.26% (135) | acyl-CoA dehydrogenase |
| M. marinum M | MMAR_5068 | fadE34 | 4e-16 | 32.87% (216) | acyl-CoA dehydrogenase FadE34 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3604c | fadE34 | 2e-15 | 31.92% (213) | acyl-CoA dehydrogenase FADE34 |
| M. gilvum PYR-GCK | Mflv_2434 | - | 3e-79 | 58.72% (281) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3573c | fadE34 | 2e-15 | 31.92% (213) | acyl-CoA dehydrogenase FADE34 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 5e-07 | 26.05% (119) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0580 | - | 2e-15 | 26.47% (272) | acyl-CoA dehydrogenase FadE |
| M. avium 104 | MAV_0900 | - | 1e-98 | 68.15% (270) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4845 | - | 1e-80 | 57.93% (271) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2022 | - | 1e-83 | 60.45% (268) | putative acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_3840 | - | 4e-16 | 35.81% (148) | acyl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4218 | - | 2e-81 | 60.99% (282) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3604c|M.bovis_AF2122/97 -------MVATVTDEQSAARELVRGWARTAASGAAATAAVRDMEYGFEEG
Rv3573c|M.tuberculosis_H37Rv -------MVATVTDEQSAARELVRGWARTAASGAAATAAVRDMEYGFEEG
MAB_0580|M.abscessus_ATCC_1997 MSVPSGLRASGSSEVQEAARDAVRQWAR----SAAVIESVRKMES-----
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 ---------MGLTSEQLELRHNARDILRAACPPDAARQAMT---------
MLBr_00737|M.leprae_Br4923 -------MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQ
Mb3604c|M.bovis_AF2122/97 NADAWRPVFAGLAGLGLFGVAVPEDCGGAGGSIEDLCAMVDEAARALVPG
Rv3573c|M.tuberculosis_H37Rv NADAWRPVFAGLAGLGLFGVAVPEDCGGAGGSIEDLCAMVDEAARALVPG
MAB_0580|M.abscessus_ATCC_1997 EPDGWGPAYVGLAELGIFGVAVPEAAGGSGAGFDDMIAMVDEAAASLVPG
MMAR_4749|M.marinum_M ---------------------------MDLTDPESLRLLADSLKTAVTQN
MAV_0900|M.avium_104 --------------------------MAPSADAESLRLLTDSLRTTMS--
MSMEG_4845|M.smegmatis_MC2_155 ------------------------------MDAESLGLLEETLRKTML--
TH_2022|M.thermoresistible__bu -------------------VSVPDISGRSDLDAESLELLEDGLRKTML--
Mflv_2434|M.gilvum_PYR-GCK --------------------MTVAIKDDAAMDEASLGMLEASLRKTMS--
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------MAPGMDEASLDMLEASLRKTML--
MUL_3840|M.ulcerans_Agy99 DPQCWRAPWKTLVDLG--WTELAASDCRDDFGSVERALVLEECGRAIAPI
MLBr_00737|M.leprae_Br4923 RARFPEEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASA
. :
Mb3604c|M.bovis_AF2122/97 PVATTA--VATLVVSDP---KLRSALASGER----FAGVAIDGGVQVDPK
Rv3573c|M.tuberculosis_H37Rv PVATTA--VATLVVSDP---KLRSALASGER----FAGVAIDGGVQVDPK
MAB_0580|M.abscessus_ATCC_1997 PVATSA--LVAVVLAADGHEELVPPLAAGER----TAGLALTGNLAIADG
MMAR_4749|M.marinum_M PSPSGA--GLDAALQALGWLDMLDEIPGTAV----PLVFAMLG--ENGVH
MAV_0900|M.avium_104 -AASGA--KLDAALTDVGWRDMLDEMPDIAI----PLVFTLLG--ETGAH
MSMEG_4845|M.smegmatis_MC2_155 -SASGA--KLDAALAELGWAEMLSDMPEVAM----PLVFRLLG--ETGSH
TH_2022|M.thermoresistible__bu -SASGA--ELDAALGELGWAQMLEDMPDTAI----PLVFRLLG--ETGSH
Mflv_2434|M.gilvum_PYR-GCK -SASGP--KLDAALTELGWADMLTEAPELAI----PLYFRLLG--ETGSH
Mvan_4218|M.vanbaalenii_PYR-1 -SSSGP--ELDAALTEMGWADMLSEVPELAV----PLVFRLLG--ETGSH
MUL_3840|M.ulcerans_Agy99 PLLSSI--DMAAGVLRGCGPAAKDVLSEIAAGVVATLAVQSPGHRLARPP
MLBr_00737|M.leprae_Br4923 SLIPAVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDA
. :
Mb3604c|M.bovis_AF2122/97 TSTASGTVGR-------------VLGGAP----GGVV---LLPADGNWLL
Rv3573c|M.tuberculosis_H37Rv TSTASGTVGR-------------VLGGAP----GGVV---LLPADGNWLL
MAB_0580|M.abscessus_ATCC_1997 --LASGVLPA-------------VLGGTA----DGVL---LAPVPDGWVL
MMAR_4749|M.marinum_M APLVNDVVAR-------------AAGCPG----GGTV--PLPFAGGSWVI
MAV_0900|M.avium_104 APLLNDVLLC-------------AAGEAP----GGTL--ALPFTGGSWVL
MSMEG_4845|M.smegmatis_MC2_155 ASVLNDVLLE-------------TIGGLP----GGTP--PMPYAGGGWVV
TH_2022|M.thermoresistible__bu ASVLNDIVLE-------------TIGALP----GGTP--PLPYAGGTWVV
Mflv_2434|M.gilvum_PYR-GCK ASILVDVVLH-------------ATGNAI----GDTVELPLPYAGNAWVV
Mvan_4218|M.vanbaalenii_PYR-1 ASVLVDVVLH-------------ATGNTI----GDTVELPLPYAGNGWVV
MUL_3840|M.ulcerans_Agy99 MVLQHGRLRG-------------HAVAVPNLARAALIVTLAATADGPVAA
MLBr_00737|M.leprae_Br4923 ASMRTRAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAF
. .
Mb3604c|M.bovis_AF2122/97 VDTACD---EVVVEPLRATDFSLPLARMVLTSAPVTVLEVSGER------
Rv3573c|M.tuberculosis_H37Rv VDTACD---EVVVEPLRATDFSLPLARMVLTSAPVTVLEVSGER------
MAB_0580|M.abscessus_ATCC_1997 VDCAEA---GVAVEAKKATDFSRPWVTVTLHGAAVRPVGLPAAV------
MMAR_4749|M.marinum_M WSRGDQ---AGS-----------VLDGELPILRVEPADPLPPA-------
MAV_0900|M.avium_104 WERRDS---CDS-----------AIDELLPIHRVPQGDPLPAA-------
MSMEG_4845|M.smegmatis_MC2_155 WERRDS---TDSA------DSANPTLGGLPLRKVPDGQLLR---------
TH_2022|M.thermoresistible__bu WERTPT---TEG------------SLGGLPLRRVPDGESMR---------
Mflv_2434|M.gilvum_PYR-GCK WDRGGA---SEATGEDRGPESLDPTLGGLPLRREEEGYPIR---------
Mvan_4218|M.vanbaalenii_PYR-1 WDRGGA---SEATGDDRSPESLDPTLGGLPLRREEEGYPIR---------
MUL_3840|M.ulcerans_Agy99 VLRRGD---AVTTRTMQSSDPAQPLADLEVDAIPALIAPVASIES-----
MLBr_00737|M.leprae_Br4923 IVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKT
:
Mb3604c|M.bovis_AF2122/97 ---VEDLAATVLAAEAAGVARWTLDTAVAYAKVREQFGKPIGSFQAVKHL
Rv3573c|M.tuberculosis_H37Rv ---VEDLAATVLAAEAAGVARWTLDTAVAYAKVREQFGKPIGSFQAVKHL
MAB_0580|M.abscessus_ATCC_1997 ---IADLSVVTLSAEAVGVARWALQTAVEYAKVREQFGKQIGSFQAIKHM
MMAR_4749|M.marinum_M ---ALAAGRSAVGWWLAGASRAMLELARRHALDRNQFGHPIAAFQAVRHR
MAV_0900|M.avium_104 ---ALHAGWRALGWWLVGTSRAMLTLGRRHAVDRVQFGRHISSFQAIRHR
MSMEG_4845|M.smegmatis_MC2_155 ----ISEARRALGWWLVGSARAMLALARQHAMDRVQFGAPIATFQAVRHR
TH_2022|M.thermoresistible__bu ----LGEARKAVGWWLVGSARAMLTLARQHALDRVQFGKPIASFQAVRHR
Mflv_2434|M.gilvum_PYR-GCK ----VAEARVAVGWWLVGSARAMLALARQHALDRVQFGKPIAGFQAVRHR
Mvan_4218|M.vanbaalenii_PYR-1 ----LAEARVAVGWWLVGSARAMLSLARQHALDRVQFGKPIASFQAVRHR
MUL_3840|M.ulcerans_Agy99 ---VVTAPLLAVSADLVGVADAVLQHAVEHAKSRHQFGVPIGGFQGIKHA
MLBr_00737|M.leprae_Br4923 ALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFM
.:. .* : * . :: * *** *. **.::.
Mb3604c|M.bovis_AF2122/97 CAQMLCRAEQADVAAADAARAAAD-----SDGTQLSIAAAVAASIGIDAA
Rv3573c|M.tuberculosis_H37Rv CAQMLCRAEQADVAAADAARAAAD-----SDGTQLSIAAAVAASIGIDAA
MAB_0580|M.abscessus_ATCC_1997 CAEMLCRVEQADVAVWDAAGCAQDVLANKGDSQQLSLAAAVAAAVAVDAA
MMAR_4749|M.marinum_M LAEALVAIEGAEATLRAATDQPDE------------LACLLAKAAAGQAA
MAV_0900|M.avium_104 LAEALVAVEGAEATLQAAAECDEDP----------GLAALLAKAAAGQAA
MSMEG_4845|M.smegmatis_MC2_155 LAETLVAIEGAEATLSQPNADNVD------------LTSLLAKAAAGKAA
TH_2022|M.thermoresistible__bu LAETLVAIEGAEAALRLPGTQSPD------------LTAMLAKAAAGKAA
Mflv_2434|M.gilvum_PYR-GCK LAETLVAIEGAEATLSLPSADNPD------------LSSLLAKAAAGKAA
Mvan_4218|M.vanbaalenii_PYR-1 LAETLVAIEGAEATLSLPGADNPD------------LTALLAKAAAGKAA
MUL_3840|M.ulcerans_Agy99 LADNYGSVERARSLTYAAASHPAP----------DWTAAALAKAAAADAA
MLBr_00737|M.leprae_Br4923 LADMAMKVEAARLIVYAAAARAERG------EPDLGFISAASKCFASDIA
*: * * . . : . . . *
Mb3604c|M.bovis_AF2122/97 KANAKDCIQVLGGIGCTWEHDAHLYLRRAHGIGGFLGGSGRWLRRVTALT
Rv3573c|M.tuberculosis_H37Rv KANAKDCIQVLGGIGCTWEHDAHLYLRRAHGIGGFLGGSGRWLRRVTALT
MAB_0580|M.abscessus_ATCC_1997 VANTKDCIQILGGIGFTWEHDAHLYLRRAYALQSFVGKPAVWRRKAAALT
MMAR_4749|M.marinum_M LTTARHCQQVLAGMGFTAEHQLHHHVKRSLVLDGLLGSSR----------
MAV_0900|M.avium_104 LTTARHCQQVLGGIGFTAEHPLHHHIKRSLILDGLLGSAR----------
MSMEG_4845|M.smegmatis_MC2_155 LTAARHCQQVLGGIGFTAEHELHTHVERVLVLDGLLGNAR----------
TH_2022|M.thermoresistible__bu LTAARNCQQVLGGIGFTAEHDLHRHVQRVLVLDGLLGSAR----------
Mflv_2434|M.gilvum_PYR-GCK LTAAKNCQQVLGGIGFTEEHGLQHHVKRTLVLDGLLGSSR----------
Mvan_4218|M.vanbaalenii_PYR-1 LTAAMHCQQVLGGIGFTEEHHLHRHVKRALVLDGLLGSSR----------
MUL_3840|M.ulcerans_Agy99 VRCATTAVQVYGAIAQTWEHHIHLYVRHAWQGAAQLGDSR----------
MLBr_00737|M.leprae_Br4923 MEVTTDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQ----------
: . *: .. . * :. . .:. *
Mb3604c|M.bovis_AF2122/97 QAGVRRRLGVDLAEVAGLRPEIAAAVAEVAALPEEKRQVALADTGLLAPH
Rv3573c|M.tuberculosis_H37Rv QAGVRRRLGVDLAEVAGLRPEIAAAVAEVAALPEEKRQVALADTGLLAPH
MAB_0580|M.abscessus_ATCC_1997 IGGIRRSLTIDLGEVESQRAEVAGQVAEIASAPKAEHQVRLAEAGFLAPH
MMAR_4749|M.marinum_M ----------ELTREAGAVLRTKGFAPRLAQL------------------
MAV_0900|M.avium_104 ----------ELTRQAGKTLVTTGSAPRLAQL------------------
MSMEG_4845|M.smegmatis_MC2_155 ----------ELTRKAGSGMRARGTAPRMAQV------------------
TH_2022|M.thermoresistible__bu ----------ELTLRVGAGLRARGSAPRLAYL------------------
Mflv_2434|M.gilvum_PYR-GCK ----------ELTRRAGAGLRARGSVPRLAQL------------------
Mvan_4218|M.vanbaalenii_PYR-1 ----------ELTRRAGAGLRARGSAPRLAQL------------------
MUL_3840|M.ulcerans_Agy99 ----------ALYHEVGRRFAGGPQ-------------------------
MLBr_00737|M.leprae_Br4923 -----------IQRVVMSRALLR---------------------------
:
Mb3604c|M.bovis_AF2122/97 WPAPYGRGASPAEQLLIDQELAAAKVERPDLVIGWWAAPTILEHGTPEQI
Rv3573c|M.tuberculosis_H37Rv WPAPYGRGASPAEQLLIDQELAAAKVERPDLVIGWWAAPTILEHGTPEQI
MAB_0580|M.abscessus_ATCC_1997 WPAPYGLGAGAAQQLLIDQELHAAGVNRPDLVIGWWAAPTILEHGTPEQI
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 ERFVPATMRGEFLWCQLFSEPGAGSDLASLRTKAVRADGGWLLTGQKVWT
Rv3573c|M.tuberculosis_H37Rv ERFVPATMRGEFLWCQLFSEPGAGSDLASLRTKAVRADGGWLLTGQKVWT
MAB_0580|M.abscessus_ATCC_1997 AQFVAPTLRGEIEWCQLFSEPGAGSDLAALRTKATRVDGGWKLDGQKVWN
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 SAAHKARWGVCLARTDPDAPKHKGITYFLVDMTTPGIEIRPLREITGDSL
Rv3573c|M.tuberculosis_H37Rv SAAHKARWGVCLARTDPDAPKHKGITYFLVDMTTPGIEIRPLREITGDSL
MAB_0580|M.abscessus_ATCC_1997 SKAQIADWAICLARTNPDVPKHKGITYFLLDMKTPGITVRPLREITGEEM
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 FNEVFLDNVFVPDEMVVGAVNDGWRLARTTLANERVAMATGTALGNPMEE
Rv3573c|M.tuberculosis_H37Rv FNEVFLDNVFVPDEMVVGAVNDGWRLARTTLANERVAMATGTALGNPMEE
MAB_0580|M.abscessus_ATCC_1997 FNEVFLDGVIVPDENVVGSVDDGWRLARTTLANERVAMAGGGTLDEAMES
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 LLKVLGDMELDVAQQDRLGRLILLAQAGALLDRRIAELAVGGQDPGAQSS
Rv3573c|M.tuberculosis_H37Rv LLKVLGDMELDVAQQDRLGRLILLAQAGALLDRRIAELAVGGQDPGAQSS
MAB_0580|M.abscessus_ATCC_1997 LLALIGDTELDAAQLDTLGAFVCSAQTGALLDLRTALRAIGGQDPGAASS
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 VRKLIGVRYRQALAEYLMEVSDGGGLVENRAVYDFLNTRCLTIAGGTEQI
Rv3573c|M.tuberculosis_H37Rv VRKLIGVRYRQALAEYLMEVSDGGGLVENRAVYDFLNTRCLTIAGGTEQI
MAB_0580|M.abscessus_ATCC_1997 VRKLVGVRHRQGLSEHILDYVDTHGAVESREMWLFMRDRCLSIAGGTTQI
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 LLTVAAERLLGLPR
Rv3573c|M.tuberculosis_H37Rv LLTVAAERLLGLPR
MAB_0580|M.abscessus_ATCC_1997 LLSVAGERLLGLPR
MMAR_4749|M.marinum_M --------------
MAV_0900|M.avium_104 --------------
MSMEG_4845|M.smegmatis_MC2_155 --------------
TH_2022|M.thermoresistible__bu --------------
Mflv_2434|M.gilvum_PYR-GCK --------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------
MUL_3840|M.ulcerans_Agy99 --------------
MLBr_00737|M.leprae_Br4923 --------------