For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDVGLTSEQLELRHNARDILRAACPPDAARQAMTDLQCWRAPWKTLVDLGWTELAASDCRDDFGSVERAL VLEECGRAIAPIPLLSSIGMAAGVLRGCGPAAKDVLSEIAAGVVATLAVQSPGHRLARPPMVLQHGRLRG HAVAVPNLTRAELIVTLAATADGPVAALLRRGDGVTTRTMQSSDPAQPLADLEVDAIPALIAPVASIESV LTAPLLAVSADLVGVADAVLQHAVEHAKSRHQFGAPIGGFQGIKHALADNYVSVERARSLTYAAASHPAP DWTAAALAKAAAADAAVRCATTAVQVYGAIAQTWEHHIHLYVRHAWQGAAQLGDSRALYHEVGRRFAGGP Q
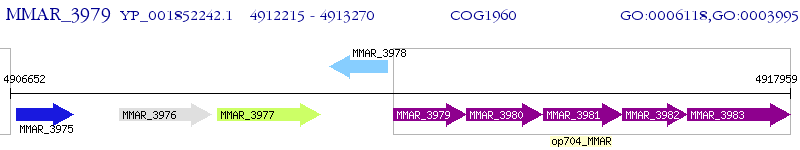
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3979 | - | - | 100% (351) | acyl-CoA dehydrogenase |
| M. marinum M | MMAR_2861 | fadE18 | 2e-33 | 32.58% (356) | acyl-CoA dehydrogenase FadE18 |
| M. marinum M | MMAR_0530 | fadE6 | 3e-31 | 32.47% (348) | acyl-CoA dehydrogenase FadE6 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1968c | fadE18 | 7e-33 | 33.15% (365) | acyl-CoA dehydrogenase FADE18 |
| M. gilvum PYR-GCK | Mflv_1795 | - | 1e-31 | 31.49% (343) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv1933c | fadE18 | 7e-33 | 33.15% (365) | acyl-CoA dehydrogenase FADE18 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 2e-16 | 27.04% (355) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4336 | - | 4e-33 | 32.20% (354) | acyl-CoA dehydrogenase FadE |
| M. avium 104 | MAV_1631 | - | 1e-124 | 66.76% (358) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5618 | - | 2e-31 | 32.39% (318) | acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2777 | - | 8e-34 | 31.83% (355) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_3840 | - | 0.0 | 97.42% (349) | acyl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4007 | - | 3e-36 | 34.45% (357) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3979|M.marinum_M ----------MDVGLTSEQLELRHNARDILRAACPP-DAARQAMTDLQCW
MUL_3840|M.ulcerans_Agy99 ------------MGLTSEQLELRHNARDILRAACPP-DAARQAMTDPQCW
MAV_1631|M.avium_104 ----------MDVGLNSEQLSLRDAVRDILRTECPP-DAARQAITDPERW
Mb1968c|M.bovis_AF2122/97 ----------MDFRYSTEQDDFRASLRGFLGRGAP---VREMAAADGSDR
Rv1933c|M.tuberculosis_H37Rv ----------MDFRYSTEQDDFRASLRGFLGRGAP---VREMAAADGSDR
Mvan_4007|M.vanbaalenii_PYR-1 ----------MDFRYHTEHDDLRAALRGFLHQTAPAARVREVAAGDGCDI
Mflv_1795|M.gilvum_PYR-GCK ----------MRAEFAEFHDELRSVASDLLAKDGH---------------
TH_2777|M.thermoresistible__bu ----------MTDQFTEFHDELRAVAAEVLAKD-R---------------
MSMEG_5618|M.smegmatis_MC2_155 ------MMTDFTTDFSAIHDELRSVAADLLAKD-----------------
MAB_4336|M.abscessus_ATCC_1997 ----------MPIAINSEHVALADSAHAFVERVVPTELLHETLETPLPWP
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAP--HAADVDQRARFP
: : .
MMAR_3979|M.marinum_M RAPWKTLVDLGWTELAASDCRDDFGS--VERALVLEECGRAIAPIPLLSS
MUL_3840|M.ulcerans_Agy99 RAPWKTLVDLGWTELAASDCRDDFGS--VERALVLEECGRAIAPIPLLSS
MAV_1631|M.avium_104 RALWKTVVDLGWTELAAANT-GDYGP--MELVVVLEECGAAIAPIPLLCS
Mb1968c|M.bovis_AF2122/97 RLWQRLCTELELPALHVPPEHGGLGATLVETAIAFAELGRALTPIP-FAA
Rv1933c|M.tuberculosis_H37Rv RLWQRLCTELELPALHVPPEHGGLGATLVETAIAFAELGRALTPIP-FAA
Mvan_4007|M.vanbaalenii_PYR-1 SLWRRLCTELELPGLHLSQECGGAGGSLVEAAIVFEELGRALTPVP-MAA
Mflv_1795|M.gilvum_PYR-GCK APEWRVLADAGWTGLEVPDTLGGAGASFAESAVILDQIGRAAGVTGYLGG
TH_2777|M.thermoresistible__bu TVDWPVLADAGWVGLEVADEFGGAGASFGEVAVICSEMGRAATASSYLGG
MSMEG_5618|M.smegmatis_MC2_155 SVDWPLLVQAGWVGLDAPESLGGAGATFAEVAVICEELGRAAATTGYLGG
MAB_4336|M.abscessus_ATCC_1997 PPFWKSAADQGLTSVHLAESVGGQGFGALELAIVVAEFGRGAVPGP-FVP
MLBr_00737|M.leprae_Br4923 EEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPA
: . .. * : : . :
MMAR_3979|M.marinum_M IG-MAAGVLRGCGP--AAKDVLSEIAAGVVATLAVQSPGHRLAR------
MUL_3840|M.ulcerans_Agy99 ID-MAAGVLRGCGP--AAKDVLSEIAAGVVATLAVQSPGHRLAR------
MAV_1631|M.avium_104 VG-LAAGLLRGSG----LDAVLADIAGGAVATVAVHAEKSRLPG------
Mb1968c|M.bovis_AF2122/97 TV-FAIEAILRMGDDEQRKRLLAGLLTGARIGTIAVSGHDVASAT---TV
Rv1933c|M.tuberculosis_H37Rv TV-FAIEAILRMGDDEQRKRLLAGLLTGARIGTIAVSGHDVASAT---TV
Mvan_4007|M.vanbaalenii_PYR-1 TV-FAIEAVLRLGDDRQRSRLLPALLSGERIATLAAARPDALDPAR-VTV
Mflv_1795|M.gilvum_PYR-GCK AV-LATGVLNLLRPSDSRDHLLAAVAGGEPRLAVVTG-------------
TH_2777|M.thermoresistible__bu AV-LGVGALNAVQPDAARDDLLTAVAAGAERTALVLG-------------
MSMEG_5618|M.smegmatis_MC2_155 AV-LGVGALLAVAPNTSRDNLLTEVVEGRTRVSLALPGDLVPGDP----R
MAB_4336|M.abscessus_ATCC_1997 SV-IASALISAHDP---DHPLLSDLASGASIATFSSTSSFTATP------
MLBr_00737|M.leprae_Br4923 VNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRT
:. : :*. :
MMAR_3979|M.marinum_M -PPMVLQHGRLRGHAVAVPNLTRAELIVTLAATADG------PVAALLRR
MUL_3840|M.ulcerans_Agy99 -PPMVLQHGRLRGHAVAVPNLARAALIVTLAATADG------PVAAVLRR
MAV_1631|M.avium_104 -APMTLRQGRLRGRAVAVPNLARAELLVTLARTDDGG-----TVAAVARC
Mb1968c|M.bovis_AF2122/97 RAVRRDGRPALTGECTPVLHGHVADLFVVPAVADGSI-----VLHVVAAD
Rv1933c|M.tuberculosis_H37Rv RAVRRDGRPALTGECTPVLHGHVADLFVVPAVADGSI-----VLHVVAAD
Mvan_4007|M.vanbaalenii_PYR-1 RADRHAGRTELSGESSPVLHGHVADLFVVPAVADGRV-----ALYVVAAD
Mflv_1795|M.gilvum_PYR-GCK -DFSMS-AGKLTGRAEFVPDAVGADRLLVVTG-----------PEVAVVE
TH_2777|M.thermoresistible__bu -PFTFT-GGRLTGRAEFVPDASGADRLLVLATDS--------DGTPVLTA
MSMEG_5618|M.smegmatis_MC2_155 VPFTVTTDGRVHGRAAFVPDAGDAHRLLLPARDA--------TGTTVLVD
MAB_4336|M.abscessus_ATCC_1997 ----AGDGLSVDGQARSVLSATSATYVVAPVAIDSD-------RRWVVLS
MLBr_00737|M.leprae_Br4923 RAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIVHKD
: * : : .
MMAR_3979|M.marinum_M GDGVTTR-TMQSSDP-AQPLADLEVDAI------PALIAPV--ASIESVL
MUL_3840|M.ulcerans_Agy99 GDAVTTR-TMQSSDP-AQPLADLEVDAI------PALIAPV--ASIESVV
MAV_1631|M.avium_104 GDGVTALPAAECTDP-AQPLAWVEIDAE------PVAVAPV--D-LESAL
Mb1968c|M.bovis_AF2122/97 APGVTVT-PLPSFDI-TRPVATLRLAGS---PAEPLTAGTP--DDMERVL
Rv1933c|M.tuberculosis_H37Rv APGVTVT-PLPSFDI-TRPVATLRLAGS---PAEPLTAGTP--DDMERVL
Mvan_4007|M.vanbaalenii_PYR-1 APGVTVD-RLPSFDT-TRPVARLRLQQA---PAEPLVAGPH--DGIERVL
Mflv_1795|M.gilvum_PYR-GCK TAAATVT-AQPVVDE-TRRLATVAADSATVVDILTFDGNPA--DAVNALR
TH_2777|M.thermoresistible__bu TAAATVT-AQPVLDE-TRRLAVVCAEDSGVEAVWRFAGDPR--AAARRLA
MSMEG_5618|M.smegmatis_MC2_155 -AAATVI-EQPVLDE-TRRLAIVEADGAEARDVWPLEDP-------HALA
MAB_4336|M.abscessus_ATCC_1997 ADDVTLD-SQQSVDP-LRPVAHVTATAVQVPADRVLSNLTD--ARATSII
MLBr_00737|M.leprae_Br4923 DEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTALATL
: . : :
MMAR_3979|M.marinum_M TAPLLAVSADLVGVADAVLQHAVEHAKSRHQFGAPIGGFQGIKHALADNY
MUL_3840|M.ulcerans_Agy99 TAPLLAVSADLVGVADAVLQHAVEHAKSRHQFGVPIGGFQGIKHALADNY
MAV_1631|M.avium_104 PAPWVACAADLVGVAGAALRRSVEHAKTRRQFGKPIGAFQGVKHALADNY
Mb1968c|M.bovis_AF2122/97 DVARVLLAAEMLGGAEACLDLAVQYAGRRTQFDRPIGSFQAVKHACADMM
Rv1933c|M.tuberculosis_H37Rv DVARVLLAAEMLGGAEACLDLAVQYAGRRTQFDRPIGSFQAVKHACADMM
Mvan_4007|M.vanbaalenii_PYR-1 DTARVLLSAEMLGGAEACLAMAVEYSCTREQFDRPIGSFQAVKHSCADMM
Mflv_1795|M.gilvum_PYR-GCK HRAEIAVACDSLGVAEQMLSDTVDYVKVRHQFGRPIGSFQAVKHACADML
TH_2777|M.thermoresistible__bu DRAAVAVAIDSLGLAEAMLSATVSYAKVRHQFGRPIGSFQAVKHACADMQ
MSMEG_5618|M.smegmatis_MC2_155 RRAALAVACDSLGIAAAMLDATVSYASMRQQFGRPIGSFQAVKHACADML
MAB_4336|M.abscessus_ATCC_1997 S---TIVSAEAVGVARWATDTASEYAKVREQFGRPIGQFQGIKHKCATMA
MLBr_00737|M.leprae_Br4923 DHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLADMA
. : :* * : : * **. .*. **.::. *
MMAR_3979|M.marinum_M VSVERARSLTYAAAS-------HPAPDWTAAALAKAAAADAAVRCATTAV
MUL_3840|M.ulcerans_Agy99 GSVERARSLTYAAAS-------HPAPDWTAAALAKAAAADAAVRCATTAV
MAV_1631|M.avium_104 VSVERARSLTYAAAARLADPGTAPGDAWTAAALAKAAAGEAATGCARTAV
Mb1968c|M.bovis_AF2122/97 IEIDATRATVMFAAMSAAN----GDELQTVAPLAKAQTAETFVLCAGSAL
Rv1933c|M.tuberculosis_H37Rv IEIDATRATVMFAAMSAAN----GDELQTVAPLAKAQTAETFVLCAGSAL
Mvan_4007|M.vanbaalenii_PYR-1 IEIDATRAAVMFAAMSAAD----PDELRITAPLVKAQAADTFTLCAGAAF
Mflv_1795|M.gilvum_PYR-GCK VAVEVSRQLVADAVDVLAG----GGDAAIPAAMAKSYVCAAAVEVAGKAM
TH_2777|M.thermoresistible__bu VQITVARQLVGRAVAALCDP--DSDDTAAAAAMAKSFVCSAAVEVVGKAM
MSMEG_5618|M.smegmatis_MC2_155 VQVTAARQLIAGAIS---------APDAHTVAKAKSYATEAAVGVAGKAM
MAB_4336|M.abscessus_ATCC_1997 AVTERATAAVWDAARALDDADENPGVVEFAASVAATLAPAAAQQCAQDCI
MLBr_00737|M.leprae_Br4923 MKVEAARLIVYAAAARAERG---EPDLGFISAASKCFASDIAMEVTTDAV
: * . . . ..
MMAR_3979|M.marinum_M QVYGAIAQTWEHHIHLYVRHAWQGAAQLGDSRALYHEVGRRFAGGPQ---
MUL_3840|M.ulcerans_Agy99 QVYGAIAQTWEHHIHLYVRHAWQGAAQLGDSRALYHEVGRRFAGGPQ---
MAV_1631|M.avium_104 QVHGALGQTWEHDAHLYVRHAWQTAALLGDSRALYHEVGRRFCGGAA---
Mb1968c|M.bovis_AF2122/97 QIHGAIAFTWEHDLHLYYRRAKTTEALFGSSARNRALLAERAGLVKA---
Rv1933c|M.tuberculosis_H37Rv QIHGAIAFTWEHDLHLYYRRAKTTEALFGSSARNRALLAERAGLVKA---
Mvan_4007|M.vanbaalenii_PYR-1 QVHGGIAFTWEHDMHLYFRRAKTTEALFGGSAHHRSLLADRVGL------
Mflv_1795|M.gilvum_PYR-GCK QLHGGIGYTWESGIHTYLKRATLNRSLFGSPAAQRRRLSQRYH-------
TH_2777|M.thermoresistible__bu QLHGGIGYTWESGVHVYLKRATLNRALYGSPAAHRSQLVSRYR-------
MSMEG_5618|M.smegmatis_MC2_155 QLHGGIGYTWESGVHTYLKRAALNRSLFGSPREHRTMLSARYRSAPG---
MAB_4336|M.abscessus_ATCC_1997 QVHGGIGYTWEHDAHIYYRRALGLTAALGRSGGAPATVVRTAVEHGLRKV
MLBr_00737|M.leprae_Br4923 QLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR------
*:.*. . * : . : : * * :
MMAR_3979|M.marinum_M --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MAV_1631|M.avium_104 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4007|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4336|M.abscessus_ATCC_1997 DIDLAPETEALRAEIRAEVAALKEIPSGKRNTAIAEGGWVLPYLPTPWGR
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MMAR_3979|M.marinum_M --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MAV_1631|M.avium_104 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4007|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4336|M.abscessus_ATCC_1997 AASPVEQVIISQEFLTGKVRRPQLGIATWLVPSIVAYGTEEQKQRFLPPT
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MMAR_3979|M.marinum_M --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MAV_1631|M.avium_104 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4007|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4336|M.abscessus_ATCC_1997 FRGEMMWCQLFSEPGAGSDLAGLSTKAVKVEGGWRLTGQKIWTSVAQFAD
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MMAR_3979|M.marinum_M --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MAV_1631|M.avium_104 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4007|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4336|M.abscessus_ATCC_1997 WGACLARTDPSAPKHNGITYFLVDMKSEGVTVRPLREMTGGALFNEVFID
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MMAR_3979|M.marinum_M --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MAV_1631|M.avium_104 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4007|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4336|M.abscessus_ATCC_1997 DVFVPDELVVGEVNRGWEVSRNTLTAERVSIGSSDLPFLASLDDLVAFVG
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MMAR_3979|M.marinum_M --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MAV_1631|M.avium_104 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4007|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4336|M.abscessus_ATCC_1997 THDLGEVARDRAGQLIAEGHGVKLLNLRSTLLTLAGGDPMPSAAVSKLLG
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MMAR_3979|M.marinum_M --------------------------------------------------
MUL_3840|M.ulcerans_Agy99 --------------------------------------------------
MAV_1631|M.avium_104 --------------------------------------------------
Mb1968c|M.bovis_AF2122/97 --------------------------------------------------
Rv1933c|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_4007|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1795|M.gilvum_PYR-GCK --------------------------------------------------
TH_2777|M.thermoresistible__bu --------------------------------------------------
MSMEG_5618|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4336|M.abscessus_ATCC_1997 MRTGQGYAEFVVNHFGQDGAIGDSEQEQGRWADYLLASRATTIYGGTSEV
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
MMAR_3979|M.marinum_M ----------------
MUL_3840|M.ulcerans_Agy99 ----------------
MAV_1631|M.avium_104 ----------------
Mb1968c|M.bovis_AF2122/97 ----------------
Rv1933c|M.tuberculosis_H37Rv ----------------
Mvan_4007|M.vanbaalenii_PYR-1 ----------------
Mflv_1795|M.gilvum_PYR-GCK ----------------
TH_2777|M.thermoresistible__bu ----------------
MSMEG_5618|M.smegmatis_MC2_155 ----------------
MAB_4336|M.abscessus_ATCC_1997 QLNIIAERLLGLPRDP
MLBr_00737|M.leprae_Br4923 ----------------