For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDAESLGLLEETLRKTMLSASGAKLDAALAELGWAEMLSDMPEVAMPLVFRLLGETGSHASVLNDVLLET IGGLPGGTPPMPYAGGGWVVWERRDSTDSADSANPTLGGLPLRKVPDGQLLRISEARRALGWWLVGSARA MLALARQHAMDRVQFGAPIATFQAVRHRLAETLVAIEGAEATLSQPNADNVDLTSLLAKAAAGKAALTAA RHCQQVLGGIGFTAEHELHTHVERVLVLDGLLGNARELTRKAGSGMRARGTAPRMAQV
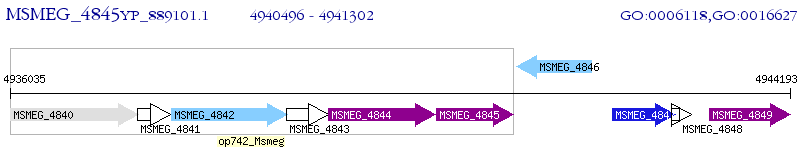
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4845 | - | - | 100% (268) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6585 | - | 8e-16 | 40.17% (117) | acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6041 | - | 2e-15 | 33.33% (183) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3604c | fadE34 | 4e-16 | 32.38% (210) | acyl-CoA dehydrogenase FADE34 |
| M. gilvum PYR-GCK | Mflv_2434 | - | 1e-105 | 69.93% (276) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3573c | fadE34 | 4e-16 | 32.38% (210) | acyl-CoA dehydrogenase FADE34 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 9e-08 | 27.68% (112) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0580 | - | 4e-17 | 28.74% (261) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_4749 | - | 9e-81 | 57.93% (271) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_0900 | - | 1e-90 | 62.50% (272) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2022 | - | 1e-116 | 76.69% (266) | putative acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_2893 | fadE18 | 9e-16 | 37.60% (125) | acyl-CoA dehydrogenase FadE18 |
| M. vanbaalenii PYR-1 | Mvan_4218 | - | 1e-108 | 71.38% (276) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3604c|M.bovis_AF2122/97 ----------MVATVTDEQSAARELVRGWARTAASGAAATAAVRDMEYGF
Rv3573c|M.tuberculosis_H37Rv ----------MVATVTDEQSAARELVRGWARTAASGAAATAAVRDMEYGF
MAB_0580|M.abscessus_ATCC_1997 ---MSVPSGLRASGSSEVQEAARDAVRQWAR----SAAVIESVRKMES--
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 ----------MDFRYSTEQDDFRESLRGFLRDQAPPARVRQVAAVGGHDK
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
Mb3604c|M.bovis_AF2122/97 EEGNADAWRPVFAGLAGLGLFG-----VAVPEDCGGAGGSIEDLCAMVDE
Rv3573c|M.tuberculosis_H37Rv EEGNADAWRPVFAGLAGLGLFG-----VAVPEDCGGAGGSIEDLCAMVDE
MAB_0580|M.abscessus_ATCC_1997 ---EPDGWGPAYVGLAELGIFG-----VAVPEAAGGSGAGFDDMIAMVDE
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------MDAESLGLLEET
TH_2022|M.thermoresistible__bu ---------------------------VSVPDISGRSDLDAESLELLEDG
Mflv_2434|M.gilvum_PYR-GCK ----------------------------MTVAIKDDAAMDEASLGMLEAS
Mvan_4218|M.vanbaalenii_PYR-1 ----------------------------------MAPGMDEASLDMLEAS
MMAR_4749|M.marinum_M -----------------------------------MDLTDPESLRLLADS
MAV_0900|M.avium_104 ----------------------------------MAPSADAESLRLLTDS
MUL_2893|M.ulcerans_Agy99 KLWHRLCAELELPALHAPPEYGGAGGTLVETAIAFGELGRALTPVPFAAT
MLBr_00737|M.leprae_Br4923 ALAALNASG--FNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPA
:
Mb3604c|M.bovis_AF2122/97 AARALVPGPVATTAV--ATLVVSDP---KLRSALASGERFAGVAIDGG--
Rv3573c|M.tuberculosis_H37Rv AARALVPGPVATTAV--ATLVVSDP---KLRSALASGERFAGVAIDGG--
MAB_0580|M.abscessus_ATCC_1997 AAASLVPGPVATSAL--VAVVLAADGHEELVPPLAAGERTAGLALTGN--
MSMEG_4845|M.smegmatis_MC2_155 LRKTML---SASGAK--LDAALAELGWAEMLSDMPEVAMPLVFRLLG---
TH_2022|M.thermoresistible__bu LRKTML---SASGAE--LDAALGELGWAQMLEDMPDTAIPLVFRLLG---
Mflv_2434|M.gilvum_PYR-GCK LRKTMS---SASGPK--LDAALTELGWADMLTEAPELAIPLYFRLLG---
Mvan_4218|M.vanbaalenii_PYR-1 LRKTML---SSSGPE--LDAALTEMGWADMLSEVPELAVPLVFRLLG---
MMAR_4749|M.marinum_M LKTAVTQNPSPSGAG--LDAALQALGWLDMLDEIPGTAVPLVFAMLG---
MAV_0900|M.avium_104 LRTTMS---AASGAK--LDAALTDVGWRDMLDEMPDIAIPLVFTLLG---
MUL_2893|M.ulcerans_Agy99 ILAIEAILRMGDSEQ--RKGLLSGLLSGDRIGALAAAGPDLSVTSVRA--
MLBr_00737|M.leprae_Br4923 VNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRT
: .
Mb3604c|M.bovis_AF2122/97 -VQVDPKTSTASGTVGRVLGG------APGGVV---LLPADGNWLLVDTA
Rv3573c|M.tuberculosis_H37Rv -VQVDPKTSTASGTVGRVLGG------APGGVV---LLPADGNWLLVDTA
MAB_0580|M.abscessus_ATCC_1997 -LAIADG--LASGVLPAVLGG------TADGVL---LAPVPDGWVLVDCA
MSMEG_4845|M.smegmatis_MC2_155 --ETGSHASVLNDVLLETIGG------LPGGTP--PMPYAGGGWVVWERR
TH_2022|M.thermoresistible__bu --ETGSHASVLNDIVLETIGA------LPGGTP--PLPYAGGTWVVWERT
Mflv_2434|M.gilvum_PYR-GCK --ETGSHASILVDVVLHATGN------AIGDTVELPLPYAGNAWVVWDRG
Mvan_4218|M.vanbaalenii_PYR-1 --ETGSHASVLVDVVLHATGN------TIGDTVELPLPYAGNGWVVWDRG
MMAR_4749|M.marinum_M --ENGVHAPLVNDVVARAAGC------PGGGTV--PLPFAGGSWVIWSRG
MAV_0900|M.avium_104 --ETGAHAPLLNDVLLCAAGE------APGGTL--ALPFTGGSWVLWERR
MUL_2893|M.ulcerans_Agy99 -GRRDGRPVLTGECAPVLHGH------VADLFVVPAESDGAVILYVVDAA
MLBr_00737|M.leprae_Br4923 RAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIVHKD
. :
Mb3604c|M.bovis_AF2122/97 CDEVVVEPLRATD--FSLPLARMVLTSAPVTVLEVSGE---------RVE
Rv3573c|M.tuberculosis_H37Rv CDEVVVEPLRATD--FSLPLARMVLTSAPVTVLEVSGE---------RVE
MAB_0580|M.abscessus_ATCC_1997 EAGVAVEAKKATD--FSRPWVTVTLHGAAVRPVGLPAA---------VIA
MSMEG_4845|M.smegmatis_MC2_155 DSTDSA------D--SANPTLGGLPLRKVPDGQLLR-------------I
TH_2022|M.thermoresistible__bu PTTEG--------------SLGGLPLRRVPDGESMR-------------L
Mflv_2434|M.gilvum_PYR-GCK GASEATGEDRGPE--SLDPTLGGLPLRREEEGYPIR-------------V
Mvan_4218|M.vanbaalenii_PYR-1 GASEATGDDRSPE--SLDPTLGGLPLRREEEGYPIR-------------L
MMAR_4749|M.marinum_M DQAGS-------------VLDGELPILRVEPADPLPPA----------AL
MAV_0900|M.avium_104 DSCDS-------------AIDELLPIHRVPQGDPLPAA----------AL
MUL_2893|M.ulcerans_Agy99 APGVAVVGLPSFD--ITRPVAKLRLVRAPAEALAASSADEV-----ARLL
MLBr_00737|M.leprae_Br4923 DEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTALATL
Mb3604c|M.bovis_AF2122/97 DLAATVLAAEAAGVARWTLDTAVAYAKVREQFGKPIGSFQAVKHLCAQML
Rv3573c|M.tuberculosis_H37Rv DLAATVLAAEAAGVARWTLDTAVAYAKVREQFGKPIGSFQAVKHLCAQML
MAB_0580|M.abscessus_ATCC_1997 DLSVVTLSAEAVGVARWALQTAVEYAKVREQFGKQIGSFQAIKHMCAEML
MSMEG_4845|M.smegmatis_MC2_155 SEARRALGWWLVGSARAMLALARQHAMDRVQFGAPIATFQAVRHRLAETL
TH_2022|M.thermoresistible__bu GEARKAVGWWLVGSARAMLTLARQHALDRVQFGKPIASFQAVRHRLAETL
Mflv_2434|M.gilvum_PYR-GCK AEARVAVGWWLVGSARAMLALARQHALDRVQFGKPIAGFQAVRHRLAETL
Mvan_4218|M.vanbaalenii_PYR-1 AEARVAVGWWLVGSARAMLSLARQHALDRVQFGKPIASFQAVRHRLAETL
MMAR_4749|M.marinum_M AAGRSAVGWWLAGASRAMLELARRHALDRNQFGHPIAAFQAVRHRLAEAL
MAV_0900|M.avium_104 HAGWRALGWWLVGTSRAMLTLGRRHAVDRVQFGRHISSFQAIRHRLAEAL
MUL_2893|M.ulcerans_Agy99 DVARVLLAAEMLGGAEACLDLAIEYACSRKQFDRPIGSFQAVKHRCADMM
MLBr_00737|M.leprae_Br4923 DHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLADMA
:. * :. * . :: * **. *. **:::. *:
Mb3604c|M.bovis_AF2122/97 CRAEQADVAAADAARAAAD-----SDGTQLSIAAAVAASIGIDAAKANAK
Rv3573c|M.tuberculosis_H37Rv CRAEQADVAAADAARAAAD-----SDGTQLSIAAAVAASIGIDAAKANAK
MAB_0580|M.abscessus_ATCC_1997 CRVEQADVAVWDAAGCAQDVLANKGDSQQLSLAAAVAAAVAVDAAVANTK
MSMEG_4845|M.smegmatis_MC2_155 VAIEGAEATLSQPNADNVD------------LTSLLAKAAAGKAALTAAR
TH_2022|M.thermoresistible__bu VAIEGAEAALRLPGTQSPD------------LTAMLAKAAAGKAALTAAR
Mflv_2434|M.gilvum_PYR-GCK VAIEGAEATLSLPSADNPD------------LSSLLAKAAAGKAALTAAK
Mvan_4218|M.vanbaalenii_PYR-1 VAIEGAEATLSLPGADNPD------------LTALLAKAAAGKAALTAAM
MMAR_4749|M.marinum_M VAIEGAEATLRAATDQPDE------------LACLLAKAAAGQAALTTAR
MAV_0900|M.avium_104 VAVEGAEATLQAAAECDEDP----------GLAALLAKAAAGQAALTTAR
MUL_2893|M.ulcerans_Agy99 IEIDATRAAVMFAQLCAAGG-------DELHATAPLAKAQAAGTYVLCAG
MLBr_00737|M.leprae_Br4923 MKVEAARLIVYAAAARAERG------EPDLGFISAASKCFASDIAMEVTT
: : . . : . . :
Mb3604c|M.bovis_AF2122/97 DCIQVLGGIGCTWEHDAHLYLRRAHGIGGFLGGSGRWLRRVTALTQAGVR
Rv3573c|M.tuberculosis_H37Rv DCIQVLGGIGCTWEHDAHLYLRRAHGIGGFLGGSGRWLRRVTALTQAGVR
MAB_0580|M.abscessus_ATCC_1997 DCIQILGGIGFTWEHDAHLYLRRAYALQSFVGKPAVWRRKAAALTIGGIR
MSMEG_4845|M.smegmatis_MC2_155 HCQQVLGGIGFTAEHELHTHVERVLVLDGLLGNAR---------------
TH_2022|M.thermoresistible__bu NCQQVLGGIGFTAEHDLHRHVQRVLVLDGLLGSAR---------------
Mflv_2434|M.gilvum_PYR-GCK NCQQVLGGIGFTEEHGLQHHVKRTLVLDGLLGSSR---------------
Mvan_4218|M.vanbaalenii_PYR-1 HCQQVLGGIGFTEEHHLHRHVKRALVLDGLLGSSR---------------
MMAR_4749|M.marinum_M HCQQVLAGMGFTAEHQLHHHVKRSLVLDGLLGSSR---------------
MAV_0900|M.avium_104 HCQQVLGGIGFTAEHPLHHHIKRSLILDGLLGSAR---------------
MUL_2893|M.ulcerans_Agy99 SAIQVHGGIAFTWEHDLHLYFRRAKTTEALFGSSA---------------
MLBr_00737|M.leprae_Br4923 DAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQ---------------
. *: .* . * :. . ... *
Mb3604c|M.bovis_AF2122/97 RRLGVDLAEVAGLRPEIAAAVAEVAALPEEKRQVALADTGLLAPHWPAPY
Rv3573c|M.tuberculosis_H37Rv RRLGVDLAEVAGLRPEIAAAVAEVAALPEEKRQVALADTGLLAPHWPAPY
MAB_0580|M.abscessus_ATCC_1997 RSLTIDLGEVESQRAEVAGQVAEIASAPKAEHQVRLAEAGFLAPHWPAPY
MSMEG_4845|M.smegmatis_MC2_155 -----ELTRKAGSGMRARGTAPRMAQV-----------------------
TH_2022|M.thermoresistible__bu -----ELTLRVGAGLRARGSAPRLAYL-----------------------
Mflv_2434|M.gilvum_PYR-GCK -----ELTRRAGAGLRARGSVPRLAQL-----------------------
Mvan_4218|M.vanbaalenii_PYR-1 -----ELTRRAGAGLRARGSAPRLAQL-----------------------
MMAR_4749|M.marinum_M -----ELTREAGAVLRTKGFAPRLAQL-----------------------
MAV_0900|M.avium_104 -----ELTRQAGKTLVTTGSAPRLAQL-----------------------
MUL_2893|M.ulcerans_Agy99 ---------------HHRSLLADRVGL-----------------------
MLBr_00737|M.leprae_Br4923 ----------------IQRVVMSRALLR----------------------
.
Mb3604c|M.bovis_AF2122/97 GRGASPAEQLLIDQELAAAKVERPDLVIGWWAAPTILEHGTPEQIERFVP
Rv3573c|M.tuberculosis_H37Rv GRGASPAEQLLIDQELAAAKVERPDLVIGWWAAPTILEHGTPEQIERFVP
MAB_0580|M.abscessus_ATCC_1997 GLGAGAAQQLLIDQELHAAGVNRPDLVIGWWAAPTILEHGTPEQIAQFVA
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 ATMRGEFLWCQLFSEPGAGSDLASLRTKAVRADGGWLLTGQKVWTSAAHK
Rv3573c|M.tuberculosis_H37Rv ATMRGEFLWCQLFSEPGAGSDLASLRTKAVRADGGWLLTGQKVWTSAAHK
MAB_0580|M.abscessus_ATCC_1997 PTLRGEIEWCQLFSEPGAGSDLAALRTKATRVDGGWKLDGQKVWNSKAQI
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 ARWGVCLARTDPDAPKHKGITYFLVDMTTPGIEIRPLREITGDSLFNEVF
Rv3573c|M.tuberculosis_H37Rv ARWGVCLARTDPDAPKHKGITYFLVDMTTPGIEIRPLREITGDSLFNEVF
MAB_0580|M.abscessus_ATCC_1997 ADWAICLARTNPDVPKHKGITYFLLDMKTPGITVRPLREITGEEMFNEVF
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 LDNVFVPDEMVVGAVNDGWRLARTTLANERVAMATGTALGNPMEELLKVL
Rv3573c|M.tuberculosis_H37Rv LDNVFVPDEMVVGAVNDGWRLARTTLANERVAMATGTALGNPMEELLKVL
MAB_0580|M.abscessus_ATCC_1997 LDGVIVPDENVVGSVDDGWRLARTTLANERVAMAGGGTLDEAMESLLALI
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 GDMELDVAQQDRLGRLILLAQAGALLDRRIAELAVGGQDPGAQSSVRKLI
Rv3573c|M.tuberculosis_H37Rv GDMELDVAQQDRLGRLILLAQAGALLDRRIAELAVGGQDPGAQSSVRKLI
MAB_0580|M.abscessus_ATCC_1997 GDTELDAAQLDTLGAFVCSAQTGALLDLRTALRAIGGQDPGAASSVRKLV
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 GVRYRQALAEYLMEVSDGGGLVENRAVYDFLNTRCLTIAGGTEQILLTVA
Rv3573c|M.tuberculosis_H37Rv GVRYRQALAEYLMEVSDGGGLVENRAVYDFLNTRCLTIAGGTEQILLTVA
MAB_0580|M.abscessus_ATCC_1997 GVRHRQGLSEHILDYVDTHGAVESREMWLFMRDRCLSIAGGTTQILLSVA
MSMEG_4845|M.smegmatis_MC2_155 --------------------------------------------------
TH_2022|M.thermoresistible__bu --------------------------------------------------
Mflv_2434|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4749|M.marinum_M --------------------------------------------------
MAV_0900|M.avium_104 --------------------------------------------------
MUL_2893|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mb3604c|M.bovis_AF2122/97 AERLLGLPR
Rv3573c|M.tuberculosis_H37Rv AERLLGLPR
MAB_0580|M.abscessus_ATCC_1997 GERLLGLPR
MSMEG_4845|M.smegmatis_MC2_155 ---------
TH_2022|M.thermoresistible__bu ---------
Mflv_2434|M.gilvum_PYR-GCK ---------
Mvan_4218|M.vanbaalenii_PYR-1 ---------
MMAR_4749|M.marinum_M ---------
MAV_0900|M.avium_104 ---------
MUL_2893|M.ulcerans_Agy99 ---------
MLBr_00737|M.leprae_Br4923 ---------