For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAIVSDQQLAARELVRDWARTAAAGEIPTAASREIERGNADAWRPVFTGLAELGLFGVAVPEDHGGAGGT IEDLCAMIEEAAKALVPGPVASTAVATAVLAEAGAEPQLLAALASGDRFAGLALEGELQFDQNTSLASGS APLVLGAIAGGVVLLPAGASWLLVDTTAEGVAVEPSAATDFSRPLARVVLAAAPALSLGELGQRVEDLTA TVLAAEAAGVTRWALETAVAYAKVREQFGKPIGSFQAIKHLCAEMLCRAEQADVAAADAARAAAESDDQQ FALAAALAAGIGISAAKANVKDCIQVLGGIGCTWEHDAHLYLRRAHSIGRFLGGAERWLRRVAALTQAGV RRRLDIDLSEVADLRPEIAAAVADVAALPEQKRQVALAEAGLLAPHWPAPYGRGASPAEQLLIDQELAAA GVVRPDLVIGWWAAPTILEHGTPEQIARFVPSTLRGDFLWCQLFSEPGAGSDLASLRTKAVRGDGGWLLT GQKVWTSAAHKARWGVCLARTDPDAPKHKGITYFLLDMTAPGIEIRPLREITGDSLFNEVFLENVFIPDQ MVVGAVNDGWRLARTTLANERVAMATGTALGNPMEELLEVLAELELDTGQQDRLARLIVATQTGALLDQR IAQLVMGGRDPGAQSSVRKLIGVRYRQALAEYMMDVSAGGGLVENRAVFDFLNTRCLTIAGGTEQILLTV AAERLLGLPR*
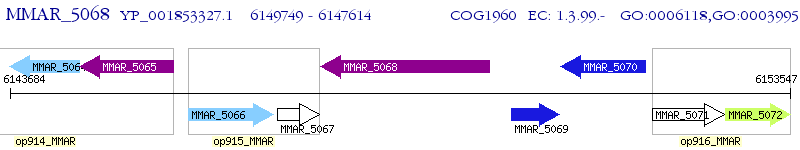
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5068 | fadE34 | - | 100% (711) | acyl-CoA dehydrogenase FadE34 |
| M. marinum M | MMAR_0530 | fadE6 | e-138 | 41.27% (693) | acyl-CoA dehydrogenase FadE6 |
| M. marinum M | MMAR_1625 | fadE22 | e-137 | 42.11% (722) | acyl-CoA dehydrogenase FadE22 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3604c | fadE34 | 0.0 | 81.12% (715) | acyl-CoA dehydrogenase FADE34 |
| M. gilvum PYR-GCK | Mflv_1460 | - | 0.0 | 69.86% (720) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3573c | fadE34 | 0.0 | 81.12% (715) | acyl-CoA dehydrogenase FADE34 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 4e-17 | 26.63% (368) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0580 | - | 0.0 | 61.32% (711) | acyl-CoA dehydrogenase FadE |
| M. avium 104 | MAV_0587 | - | 0.0 | 81.72% (711) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6041 | - | 0.0 | 69.41% (716) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_1744 | fadE34 | 0.0 | 72.73% (682) | PROBABLE ACYL-CoA DEHYDROGENASE FADE34 |
| M. ulcerans Agy99 | MUL_4144 | fadE34 | 0.0 | 99.02% (711) | acyl-CoA dehydrogenase FadE34 |
| M. vanbaalenii PYR-1 | Mvan_5311 | - | 0.0 | 72.28% (718) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1460|M.gilvum_PYR-GCK ---MPVPTSATITDEQADARELVRSWATASGS----VEAAREVEQ----G
Mvan_5311|M.vanbaalenii_PYR-1 ---MSASTSAPTTDDQSAARDLVRTWAAASRA----VEAARDVEQ----G
MSMEG_6041|M.smegmatis_MC2_155 ---MSLLSAGAGTSEEFAARDMVRSWAAASGA----VAAVRESE-----T
TH_1744|M.thermoresistible__bu ------------------------------------VSAVREVER----G
Mb3604c|M.bovis_AF2122/97 -------MVATVTDEQSAARELVRGWARTAASGAAATAAVRDMEYGFEEG
Rv3573c|M.tuberculosis_H37Rv -------MVATVTDEQSAARELVRGWARTAASGAAATAAVRDMEYGFEEG
MMAR_5068|M.marinum_M -------MVAIVSDQQLAARELVRDWARTAAAGEIPTAASREIER----G
MUL_4144|M.ulcerans_Agy99 -------MVAIVSDQQLAARELVRDWARTAAAGEIPTAASREIER----G
MAV_0587|M.avium_104 -------MVATVTDEQFAARALVRDWARNASTGPGGTAAIRDVEQ----G
MAB_0580|M.abscessus_ATCC_1997 MSVPSGLRASGSSEVQEAARDAVRQWARSAAV----IESVRKMES-----
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1460|M.gilvum_PYR-GCK DAAAWQAPYGRLAELGIFGVALPEAHGGADGTVADLAAMVDEAAAALVPG
Mvan_5311|M.vanbaalenii_PYR-1 DPDAWRAPYAGLAELGIFGVALPEEHGGADGTVSDLAAMIDEAAAALVPG
MSMEG_6041|M.smegmatis_MC2_155 DPGAWRAAYQGFAELGIFGVAVPDELGGAGSTVADLAAMIDEAAAALVPG
TH_1744|M.thermoresistible__bu RPDSWRGPFAGFAELGTFGVAVAEEHGGAGGTIADLCAMVDEAAAALVPG
Mb3604c|M.bovis_AF2122/97 NADAWRPVFAGLAGLGLFGVAVPEDCGGAGGSIEDLCAMVDEAARALVPG
Rv3573c|M.tuberculosis_H37Rv NADAWRPVFAGLAGLGLFGVAVPEDCGGAGGSIEDLCAMVDEAARALVPG
MMAR_5068|M.marinum_M NADAWRPVFTGLAELGLFGVAVPEDHGGAGGTIEDLCAMIEEAAKALVPG
MUL_4144|M.ulcerans_Agy99 NADAWRPVFTGLAELGLFGVAVPEDHGGAGGIIEDLCAMIEEAAKALVPG
MAV_0587|M.avium_104 NPDAWRPVFARLAELGIFGVAVPEEAGGAGGSVEDLCAMVEEAAKALIPG
MAB_0580|M.abscessus_ATCC_1997 EPDGWGPAYVGLAELGIFGVAVPEAAGGSGAGFDDMIAMVDEAAASLVPG
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1460|M.gilvum_PYR-GCK PVATTALATLVLPDG--QADLLESLATGQRWAGVALNADLSYADGR--VS
Mvan_5311|M.vanbaalenii_PYR-1 PVATTALATLVLSGP--HADLLEELATGERTAGVTLTADLDYADGR--VS
MSMEG_6041|M.smegmatis_MC2_155 PVVTTALATLLVTD----TGLLEALMSGERTAGVALSADVTVVDGR--AS
TH_1744|M.thermoresistible__bu PVATTALATLTVSD----PELLEAFAAGQRTAGVALGGEVRFADGR--AS
Mb3604c|M.bovis_AF2122/97 PVATTAVATLVVSDP----KLRSALASGERFAGVAIDGGVQVDPKTSTAS
Rv3573c|M.tuberculosis_H37Rv PVATTAVATLVVSDP----KLRSALASGERFAGVAIDGGVQVDPKTSTAS
MMAR_5068|M.marinum_M PVASTAVATAVLAEAGAEPQLLAALASGDRFAGLALEGELQFDQNTSLAS
MUL_4144|M.ulcerans_Agy99 PVASTAVATAVLAEAGAEPQLLAALASGDRFAGLALEGELQFDQNTSLAS
MAV_0587|M.avium_104 PVATTALATLVLSDA----ELLAALAAGERFAGLALQGDITFDGAASTAS
MAB_0580|M.abscessus_ATCC_1997 PVATSALVAVVLAADG-HEELVPPLAAGERTAGLALTGNLAIADGL--AS
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1460|M.gilvum_PYR-GCK GTAAYVLGADESAVLLLPVGET-WLLVDAGADGVTVEPLQATDFSRPLAR
Mvan_5311|M.vanbaalenii_PYR-1 GSAPYVLGAAATGVLLLPAGDT-WLLVDAAADGVTVESLKATDFSRPLAR
MSMEG_6041|M.smegmatis_MC2_155 GTVEFVLGADAAGVLLLPAGDD-VLLLDAGADGVSVEPLTATDFSIPLAR
TH_1744|M.thermoresistible__bu GTVPYVLGADPAGVLLLPAGTAGWVLVDAAADGVTLEPLEATDFSRPLAR
Mb3604c|M.bovis_AF2122/97 GTVGRVLGGAPGGVVLLPADGN-WLLVDTACDEVVVEPLRATDFSLPLAR
Rv3573c|M.tuberculosis_H37Rv GTVGRVLGGAPGGVVLLPADGN-WLLVDTACDEVVVEPLRATDFSLPLAR
MMAR_5068|M.marinum_M GSAPLVLGAIAGGVVLLPAGAS-WLLVDTTAEGVAVEPSAATDFSRPLAR
MUL_4144|M.ulcerans_Agy99 GSAPLVLGAIAGGVVLLPAGAS-WLLVDTTAEGVAVEPLAATDFSRPLAR
MAV_0587|M.avium_104 GTLPFVLGAADGGVLLLPADGR-WLVVDTASDGVHIEPLAATDFSRPLAR
MAB_0580|M.abscessus_ATCC_1997 GVLPAVLGGTADGVLLAPVPDG-WVLVDCAEAGVAVEAKKATDFSRPWVT
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1460|M.gilvum_PYR-GCK VSFDGAPAEALPVSGQRVVDLAATVMAAESAGLARWLLHTATEYAKVREQ
Mvan_5311|M.vanbaalenii_PYR-1 VVLDGAPAEVLDVPSQQVVDMAATLLAAEAAGLARWLLDTATEYAKVREQ
MSMEG_6041|M.smegmatis_MC2_155 VELDNAPAEVLPVSRQRFSDLAATLLAAEAAGLARWTLNTATEYAKVREQ
TH_1744|M.thermoresistible__bu VSLDSVAAQQLSIPSQRFEDLAATVLAAEAAGISRWLLHTATEYAKVREQ
Mb3604c|M.bovis_AF2122/97 MVLTSAPVTVLEVSGERVEDLAATVLAAEAAGVARWTLDTAVAYAKVREQ
Rv3573c|M.tuberculosis_H37Rv MVLTSAPVTVLEVSGERVEDLAATVLAAEAAGVARWTLDTAVAYAKVREQ
MMAR_5068|M.marinum_M VVLAAAPALSLGELGQRVEDLTATVLAAEAAGVTRWALETAVAYAKVREQ
MUL_4144|M.ulcerans_Agy99 VVLAAAPALSLGELGQRVEDLTATVLAAEAAGVTRWALETAVAYAKVREQ
MAV_0587|M.avium_104 VVLDSAPATVLAQTPERVEELAATVLAAEAAGITRWSLQTAVDYAKVREQ
MAB_0580|M.abscessus_ATCC_1997 VTLHGAAVRPVGLPAAVIADLSVVTLSAEAVGVARWALQTAVEYAKVREQ
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1460|M.gilvum_PYR-GCK FGKPIGSFQAIKHMCAEMLLRSEQASVAAADAARAASDPAADG---AQLS
Mvan_5311|M.vanbaalenii_PYR-1 FGKPIGSFQAIKHMCAEMLLRSEQASVAAADAARAAGESDAD-----QLS
MSMEG_6041|M.smegmatis_MC2_155 FGKPIGSFQAIKHMCAEMLLRSEQIAVAAADAAKAVAESDGSDS-ATQLS
TH_1744|M.thermoresistible__bu FGQPIGRFQAVKHMCAEMLLRSEQISVAAADAARAAAEYDDD-----QLS
Mb3604c|M.bovis_AF2122/97 FGKPIGSFQAVKHLCAQMLCRAEQADVAAADAARAAADSDGT-----QLS
Rv3573c|M.tuberculosis_H37Rv FGKPIGSFQAVKHLCAQMLCRAEQADVAAADAARAAADSDGT-----QLS
MMAR_5068|M.marinum_M FGKPIGSFQAIKHLCAEMLCRAEQADVAAADAARAAAESDDQ-----QFA
MUL_4144|M.ulcerans_Agy99 FGKPIGSFQAIKHLCAEMLCRAEQADVAAADAARAAAESDDQ-----QFA
MAV_0587|M.avium_104 FGKPIGSFQAIKHLCAEMLCRAEQAEVAAADAARAAAEDDPA-----QFS
MAB_0580|M.abscessus_ATCC_1997 FGKQIGSFQAIKHMCAEMLCRVEQADVAVWDAAGCAQDVLANKGDSQQLS
MLBr_00737|M.leprae_Br4923 --------------------------------------------------
Mflv_1460|M.gilvum_PYR-GCK IAAALAASVGIEAAKANAKDCIQVLGGIGITWEHDAHLYLRRAYGIAQFL
Mvan_5311|M.vanbaalenii_PYR-1 IAAALAASVGIEAAKANAKDCIQVLGGIGITWEHDAHLYLRRAYGIAQFL
MSMEG_6041|M.smegmatis_MC2_155 IAAAVAVATGIDAAKANAKDCIQVLGGIGITWEHDAHLYLRRAYGIAHFL
TH_1744|M.thermoresistible__bu IAAAVAAAAGIDAAQANSKDCIQVLGGIGITWEHDAHLYLRRAYGIAQFL
Mb3604c|M.bovis_AF2122/97 IAAAVAASIGIDAAKANAKDCIQVLGGIGCTWEHDAHLYLRRAHGIGGFL
Rv3573c|M.tuberculosis_H37Rv IAAAVAASIGIDAAKANAKDCIQVLGGIGCTWEHDAHLYLRRAHGIGGFL
MMAR_5068|M.marinum_M LAAALAAGIGISAAKANVKDCIQVLGGIGCTWEHDAHLYLRRAHSIGRFL
MUL_4144|M.ulcerans_Agy99 LAAALAASIGISAAKANVKDCIQVLGGIGCTWEHDAHLYLRRAHSIGRFL
MAV_0587|M.avium_104 LAAALAAATAITAVKANVKDCIQVLGGIGCTWEHDAHLYLRRAHAIGRFL
MAB_0580|M.abscessus_ATCC_1997 LAAAVAAAVAVDAAVANTKDCIQILGGIGFTWEHDAHLYLRRAYALQSFV
MLBr_00737|M.leprae_Br4923 ---------------------------------------------MVGWS
: :
Mflv_1460|M.gilvum_PYR-GCK GGRSHWLRRVAALTQQGVRRELHIDLESVADLRPEIASAVAEVAAAPEDG
Mvan_5311|M.vanbaalenii_PYR-1 GGRSRWLRRVAALTQQGVRRELHIDLESVAGLRPEIVSAVAEVAAAPEDK
MSMEG_6041|M.smegmatis_MC2_155 GGSRRWVRRVAALTRQGARRDIDIDLESVADLRPEISAAVAEVAALPEEK
TH_1744|M.thermoresistible__bu GGRPRWLRRVAALTQRGVRRELRIDLESVEHLRPEIRSLVADVAALPVEK
Mb3604c|M.bovis_AF2122/97 GGSGRWLRRVTALTQAGVRRRLGVDLAEVAGLRPEIAAAVAEVAALPEEK
Rv3573c|M.tuberculosis_H37Rv GGSGRWLRRVTALTQAGVRRRLGVDLAEVAGLRPEIAAAVAEVAALPEEK
MMAR_5068|M.marinum_M GGAERWLRRVAALTQAGVRRRLDIDLSEVADLRPEIAAAVADVAALPEQK
MUL_4144|M.ulcerans_Agy99 GGAERWLRRVAALTQAGVRRRLDIDLSEVADLRPEIVAAVADVAALPEQK
MAV_0587|M.avium_104 GGAERWLRRITALTQAGVRRRLGIDLTEVEDQRPQIAEAVARIAALPAEQ
MAB_0580|M.abscessus_ATCC_1997 GKPAVWRRKAAALTIGGIRRSLTIDLGEVESQRAEVAGQVAEIASAPKAE
MLBr_00737|M.leprae_Br4923 GNP-----LFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
* :. :. : : : . . :
Mflv_1460|M.gilvum_PYR-GCK RQVALAETGLLAPHWPKPYG--RAAGPAEQLLIDQELASAGVVRPDLVIG
Mvan_5311|M.vanbaalenii_PYR-1 RQVALAEARLLAPHWPPPHG--RAAGPAEQLLIDQELAAAGVVRPDLVIG
MSMEG_6041|M.smegmatis_MC2_155 RQVALAESGLLAPHWPKPYG--RNASPAEQLLIDQELAAAEVERPDLVIG
TH_1744|M.thermoresistible__bu RQAALADAGLQAPHWPRPYG--RDASPAEQLLIDQELAAAGVERPDLVIG
Mb3604c|M.bovis_AF2122/97 RQVALADTGLLAPHWPAPYG--RGASPAEQLLIDQELAAAKVERPDLVIG
Rv3573c|M.tuberculosis_H37Rv RQVALADTGLLAPHWPAPYG--RGASPAEQLLIDQELAAAKVERPDLVIG
MMAR_5068|M.marinum_M RQVALAEAGLLAPHWPAPYG--RGASPAEQLLIDQELAAAGVVRPDLVIG
MUL_4144|M.ulcerans_Agy99 RQEALAEAGLLAPHWPAPYG--RGASPAEQLLIDQELAAAGVVRPDLVIG
MAV_0587|M.avium_104 RQAALAEAGLQAPHWPAPYG--RGASPAEQLLIDQELAAAGVARPDLVIG
MAB_0580|M.abscessus_ATCC_1997 HQVRLAEAGFLAPHWPAPYG--LGAGAAQQLLIDQELHAAGVNRPDLVIG
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
* : : * * * :* . * ::*:: .:.
Mflv_1460|M.gilvum_PYR-GCK WWAVPTILEHGSPELIEQFVPPTLRGELTWCQLFSEPGAGSDLAALRMKA
Mvan_5311|M.vanbaalenii_PYR-1 WWAVPTILEHGSPEQVEQFVPPTLRGELTWCQLFSEPGAGSDLAALRTKA
MSMEG_6041|M.smegmatis_MC2_155 WWAVPTILEHGSPEQIEQFVPATLRGDLFWCQLFSEPGAGSDLAALRTKA
TH_1744|M.thermoresistible__bu WWAVPTILSHGSPEQIERFVSPTLRGELEWCQLFSEPGAGSDLAALRTKA
Mb3604c|M.bovis_AF2122/97 WWAAPTILEHGTPEQIERFVPATMRGEFLWCQLFSEPGAGSDLASLRTKA
Rv3573c|M.tuberculosis_H37Rv WWAAPTILEHGTPEQIERFVPATMRGEFLWCQLFSEPGAGSDLASLRTKA
MMAR_5068|M.marinum_M WWAAPTILEHGTPEQIARFVPSTLRGDFLWCQLFSEPGAGSDLASLRTKA
MUL_4144|M.ulcerans_Agy99 WWAAPTILEHGTPEQIARFVPSTLRGDFLWCQLFSEPGAGSDLASLRTKA
MAV_0587|M.avium_104 WWAAPTILEHGTPEQIERFVPGTLRGEFLWCQLFSEPGAGSDLASLRTKA
MAB_0580|M.abscessus_ATCC_1997 WWAAPTILEHGTPEQIAQFVAPTLRGEIEWCQLFSEPGAGSDLAALRTKA
MLBr_00737|M.leprae_Br4923 KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRTRA
.. :: :*: * :.:. . :** **** *::* :*
Mflv_1460|M.gilvum_PYR-GCK VRAEGASASGAKVSGWKLTGQKVWTSAAQKAHWGVCLARTDPEAAKHKGI
Mvan_5311|M.vanbaalenii_PYR-1 VRVDDASASGASGPGWKLTGQKVWTSAAQKAHWGVCLARTDPDAPKHKGI
MSMEG_6041|M.smegmatis_MC2_155 VRAEGPNGE----PGWKLTGQKVWTSAAQNAHWGVCLARTDPDAPKHKGI
TH_1744|M.thermoresistible__bu VRVDG---------GWRLTGQKVWTSAAHKADWGVCLARTDPDAPKHKGI
Mb3604c|M.bovis_AF2122/97 VRADG---------GWLLTGQKVWTSAAHKARWGVCLARTDPDAPKHKGI
Rv3573c|M.tuberculosis_H37Rv VRADG---------GWLLTGQKVWTSAAHKARWGVCLARTDPDAPKHKGI
MMAR_5068|M.marinum_M VRGDG---------GWLLTGQKVWTSAAHKARWGVCLARTDPDAPKHKGI
MUL_4144|M.ulcerans_Agy99 VRGDG---------GWLLTGQKVWTSAAHKARWGVCLARTDPDAPKHKGI
MAV_0587|M.avium_104 VRTDG---------GWLLTGQKVWTSAAHKARWGVCLARTDPDAPKHKGI
MAB_0580|M.abscessus_ATCC_1997 TRVDG---------GWKLDGQKVWNSKAQIADWAICLARTNPDVPKHKGI
MLBr_00737|M.leprae_Br4923 KADGD---------DWILNGFKCWITNGGKSTWYTVMAVTDPDK-GANGI
. .* * * * * : . : * :* *:*: :**
Mflv_1460|M.gilvum_PYR-GCK TYFLIDMKSPGIVIRPLREITG--DELFNEVFFDDVFVPDEMVVGEVNQG
Mvan_5311|M.vanbaalenii_PYR-1 TYFLIDMKSPGIVIRPLREITG--DELFNEVFFDDVFVPDEMVVGQVNDG
MSMEG_6041|M.smegmatis_MC2_155 TYFLVDMKTPGIVIRPLREITG--DELFNEVFFDDVFVPDTMVVGTVNDG
TH_1744|M.thermoresistible__bu TYFLVDMKSPGIEIRPLREITG--DELFNEVFFDEVFVPDEMVVGQVNDG
Mb3604c|M.bovis_AF2122/97 TYFLVDMTTPGIEIRPLREITG--DSLFNEVFLDNVFVPDEMVVGAVNDG
Rv3573c|M.tuberculosis_H37Rv TYFLVDMTTPGIEIRPLREITG--DSLFNEVFLDNVFVPDEMVVGAVNDG
MMAR_5068|M.marinum_M TYFLLDMTAPGIEIRPLREITG--DSLFNEVFLENVFIPDQMVVGAVNDG
MUL_4144|M.ulcerans_Agy99 TYFLLDMTAPGIEIRPLREITG--DSLFNEVFLENVFIPDQMVVGAVNDG
MAV_0587|M.avium_104 TYFLVDMSAPGIEIRPLREITG--DSLFNEVFLDNVFVPDEMVVGEVNDG
MAB_0580|M.abscessus_ATCC_1997 TYFLLDMKTPGITVRPLREITG--EEMFNEVFLDGVIVPDENVVGSVDDG
MLBr_00737|M.leprae_Br4923 SAFIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTG
: *::. *: : * .: * . .*:::: :*. ::* . *
Mflv_1460|M.gilvum_PYR-GCK WRLARTTLANER---VAMAQGTALGNPMEELLR------VIAELDVDGAQ
Mvan_5311|M.vanbaalenii_PYR-1 WRLARTTLANER---VAMAQGTALGNPMEELLR------VVAQLEVDGAQ
MSMEG_6041|M.smegmatis_MC2_155 WRLARTTLANER---VAMAGGTALGNPMEEVLRA-----VEAQPEFDPAD
TH_1744|M.thermoresistible__bu WRLARTTLANER---VAMAQGTALGNPMEELLR------TVTDLELDVAE
Mb3604c|M.bovis_AF2122/97 WRLARTTLANER---VAMATGTALGNPMEELLK------VLGDMELDVAQ
Rv3573c|M.tuberculosis_H37Rv WRLARTTLANER---VAMATGTALGNPMEELLK------VLGDMELDVAQ
MMAR_5068|M.marinum_M WRLARTTLANER---VAMATGTALGNPMEELLE------VLAELELDTGQ
MUL_4144|M.ulcerans_Agy99 WRLARTTLANER---VAMATGTALGNPMEELLE------VLAEPELDTGQ
MAV_0587|M.avium_104 WRLARTTLANER---VAMASGTALGNPMEELLE------LLSTLPLDSAQ
MAB_0580|M.abscessus_ATCC_1997 WRLARTTLANER---VAMAGGGTLDEAMESLLA------LIGDTELDAAQ
MLBr_00737|M.leprae_Br4923 FKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSI
:: * :** : * * * * : . :: .
Mflv_1460|M.gilvum_PYR-GCK QDRLGHLIGTAQVGTLLDQRIAQLAVEGQ-DPGPQASARKLIGVRYRQAL
Mvan_5311|M.vanbaalenii_PYR-1 QDQLGQLIVTAQVGSLLDQRIAQLAVEGQ-DPGPQASARKLIGVRYRQAL
MSMEG_6041|M.smegmatis_MC2_155 QDRLGALIVTAQVGSLLDQRIAQLAVSGQ-DTGAQASARKLIGVRYRQAL
TH_1744|M.thermoresistible__bu QDRLGGLIVSAQVGSLLDQRIAQKAVGGQ-DPGAESSARKLIGVRHRQEL
Mb3604c|M.bovis_AF2122/97 QDRLGRLILLAQAGALLDRRIAELAVGGQ-DPGAQSSVRKLIGVRYRQAL
Rv3573c|M.tuberculosis_H37Rv QDRLGRLILLAQAGALLDRRIAELAVGGQ-DPGAQSSVRKLIGVRYRQAL
MMAR_5068|M.marinum_M QDRLARLIVATQTGALLDQRIAQLVMGGR-DPGAQSSVRKLIGVRYRQAL
MUL_4144|M.ulcerans_Agy99 QDRLARLIVATQTGALLDQRIAQLVMGGR-DPGAQSSVRKLIGVRYRQAL
MAV_0587|M.avium_104 QDRLGRLIVTAQTGALLDQRIAQLAVGGQ-DPGAQSSVRKLIGVRYRQAL
MAB_0580|M.abscessus_ATCC_1997 LDTLGAFVCSAQTGALLDLRTALRAIGGQ-DPGAASSVRKLVGVRHRQGL
MLBr_00737|M.leprae_Br4923 QFMLADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEV
*. : .:.. *: * . *. * * ::. * .. :
Mflv_1460|M.gilvum_PYR-GCK AEFRMELSEGAGAIVNQQVHDFLN-TRCLTIAGGTEQILLTMAGERLLGL
Mvan_5311|M.vanbaalenii_PYR-1 AEFRMDLSDGAGAVENRQVHDFLN-TRCLTIAGGTEQILLTMAGERLLGL
MSMEG_6041|M.smegmatis_MC2_155 AEFHMELTEGGGAVRNREVHTFLN-TRCLTIAGGTEQILLTLAGERLLGL
TH_1744|M.thermoresistible__bu SEFRMELSDGAGVVHNPTVHTFLN-ARCLTIAGGTEQILLTLAGERLLGL
Mb3604c|M.bovis_AF2122/97 AEYLMEVSDGGGLVENRAVYDFLN-TRCLTIAGGTEQILLTVAAERLLGL
Rv3573c|M.tuberculosis_H37Rv AEYLMEVSDGGGLVENRAVYDFLN-TRCLTIAGGTEQILLTVAAERLLGL
MMAR_5068|M.marinum_M AEYMMDVSAGGGLVENRAVFDFLN-TRCLTIAGGTEQILLTVAAERLLGL
MUL_4144|M.ulcerans_Agy99 AEYMMDVSACGGLVENRAVFDFLN-TRCLTIAGGTEQILLTVAAERLLGL
MAV_0587|M.avium_104 AEYTMDVSEGGGLVHNRAVYDFLN-TRCLTIAGGTEQILLTVAAERLLGL
MAB_0580|M.abscessus_ATCC_1997 SEHILDYVDTHGAVESREMWLFMR-DRCLSIAGGTTQILLSVAGERLLGL
MLBr_00737|M.leprae_Br4923 TTDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR-
: :: * . : *:. : * ** ** : .. **
Mflv_1460|M.gilvum_PYR-GCK PR
Mvan_5311|M.vanbaalenii_PYR-1 PR
MSMEG_6041|M.smegmatis_MC2_155 PR
TH_1744|M.thermoresistible__bu PR
Mb3604c|M.bovis_AF2122/97 PR
Rv3573c|M.tuberculosis_H37Rv PR
MMAR_5068|M.marinum_M PR
MUL_4144|M.ulcerans_Agy99 PR
MAV_0587|M.avium_104 PR
MAB_0580|M.abscessus_ATCC_1997 PR
MLBr_00737|M.leprae_Br4923 --