For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MALWGDGISALLIDGKLSDGRAGTFPTVNPATEEVLGVAADADAEDMGRAIEAARRAFDSTDWSRNTELR VRCVRQLRDAMQQHVEELRELTISEVGAPRMLTASAQLEGPVGDLSFAADTAESYPWKQDLGEASPLGIA TRRTLAREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTPWCAAALGEIIVEHTDFPPGVV NIVTSSSHALGALLAKDPRVDMISFTGSTATGRAVMADAAATIKKVFLELGGKSAFVVLDDADLAAASAV SAFSACMHAGQGCAITTRLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVCGPLISARQRDRVQGYLDL AVAEGGRFACGGARPADREVGFYIEPTVIAGLTNDARVAREEIFGPVLTVIAHDGDDDAVRIANDSPYGL SGTVYGADPQRAARIASRLRVGTVNVNGGVWYCADAPFGGYKQSGIGREMGLLGFEEYLEAKLIATAAN
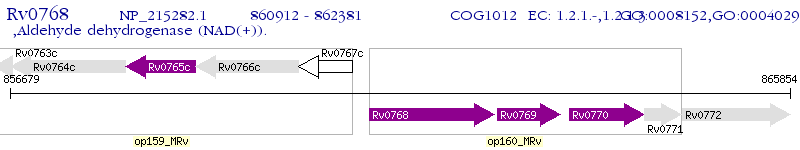
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0768 | aldA | - | 100% (489) | PROBABLE ALDEHYDE DEHYDROGENASE NAD DEPENDENT ALDA (ALDEHYDE DEHYDROGENASE |
| M. tuberculosis H37Rv | Rv0223c | - | 2e-95 | 41.48% (458) | aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv2858c | aldC | 5e-72 | 38.12% (467) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0791 | aldA | 0.0 | 100.00% (489) | aldehyde dehydrogenase NAD dependant AldA |
| M. gilvum PYR-GCK | Mflv_1600 | - | 0.0 | 79.35% (489) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 1e-60 | 33.33% (459) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1218 | - | 0.0 | 79.18% (490) | aldehyde dehydrogenase AldA |
| M. marinum M | MMAR_4928 | aldA | 0.0 | 86.09% (489) | NAD-dependent aldehyde dehydrogenase, AldA |
| M. avium 104 | MAV_0713 | - | 0.0 | 83.78% (487) | aldehyde dehydrogenase (NAD) family protein |
| M. smegmatis MC2 155 | MSMEG_5859 | - | 0.0 | 79.71% (488) | aldehyde dehydrogenase (NAD) family protein |
| M. thermoresistible (build 8) | TH_2814 | - | 0.0 | 79.88% (487) | PUTATIVE aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_0477 | aldA | 0.0 | 85.28% (489) | NAD-dependent aldehyde dehydrogenase, AldA |
| M. vanbaalenii PYR-1 | Mvan_5157 | - | 0.0 | 79.96% (489) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0768|M.tuberculosis_H37Rv MALWGDGISALLIDGKLSDGRAGTFPTVNPATEEVLGVAADADAEDMGRA
Mb0791|M.bovis_AF2122/97 MALWGDGISALLIDGKLSDGRAGTFPTVNPATEEVLGVAADADAEDMGRA
MMAR_4928|M.marinum_M MALLADGVSSLFIDGKRCEGAAGTFATVNPATEETLGVAADADAGDMDRA
MUL_0477|M.ulcerans_Agy99 MALLADGVSSLFIDGKRCEGAAGTFATVNPATEETLGVAADAVAGDMDRA
MAV_0713|M.avium_104 MALLADGVSELFIDGKMSAGNAGTFPTVNPATEEVLGVAANADAADMDRA
Mflv_1600|M.gilvum_PYR-GCK MAILADRDSKLLIDGKLVAGSAGTFPTVNPATEETLGVAADAGADDMDAA
Mvan_5157|M.vanbaalenii_PYR-1 MAILADRESRLLIDGKLVAGSSGTFTTVNPATEETLGVAADASAEDMSAA
MSMEG_5859|M.smegmatis_MC2_155 MPLLADRQSQLLIDGKLVAGRKGVFDTVNPATEEVLGVAADATAEDMGDA
TH_2814|M.thermoresistible__bu MSLLADRESRLLIDGKLVAGSAGAFTTINPATEDVLGVAANADADDMVRA
MAB_1218|M.abscessus_ATCC_1997 MTVLSDGDSELFIDGKLLPGGAGRFPTVNPATEEVLGTAADADSDDMSDA
MLBr_02573|M.leprae_Br4923 ----------------------MSIATINPATGETVKTFTPTTQDEVEDA
: *:**** :.: . : : :: *
Rv0768|M.tuberculosis_H37Rv IEAARRAFDSTDWSRNTELRVRCVRQLRDAMQQHVEELRELTISEVGAPR
Mb0791|M.bovis_AF2122/97 IEAARRAFDSTDWSRNTELRVRCVRQLRDAMQQHVEELRELTISEVGAPR
MMAR_4928|M.marinum_M IEAARRAFDDTDWSRNTELRVRCVRQLRDAMREHIEELRALTISEVGAPR
MUL_0477|M.ulcerans_Agy99 IEAARRAFDDTDWSRNTELRVRCVRQLRDAMREHIEELRALTISEVGAPR
MAV_0713|M.avium_104 IDAARRAFDETDWSRNTELRVRCVRQLRDAMREHIEELRDITIAEVGAPR
Mflv_1600|M.gilvum_PYR-GCK IGAARRAFDETGWSTDTALRVRCIRQLQNAMRAHVEELRELTIAEVGAPR
Mvan_5157|M.vanbaalenii_PYR-1 IGAARRSFDETDWATNTDLRVRCIRQLQQAMRDHVEELRELTIAEVGAPR
MSMEG_5859|M.smegmatis_MC2_155 IEAARRAFDDTDWSTDTELRVRCIRQLRDALREHVEELRDLTIAEVGAPR
TH_2814|M.thermoresistible__bu IDAARRAFDDTDWSTNTALRVRCLRQLQQAMRDHLEELRELTIAEVGAPR
MAB_1218|M.abscessus_ATCC_1997 IEAARRAFDDSGWSRDVALRVHCIRQLRDGMKTHIEELRELTIAEVGAPR
MLBr_02573|M.leprae_Br4923 IARAYARFDN-YRHTTFTQRAKWANATADLLEAEANQSAALMTLEMGKT-
* * **. *.: . : :. . :: : *:* .
Rv0768|M.tuberculosis_H37Rv MLTASAQLEGPVGDLSFAADTAESYPWKQDLGEASPLGIATRRTLAREAV
Mb0791|M.bovis_AF2122/97 MLTASAQLEGPVGDLSFAADTAESYPWKQDLGEASPLGIATRRTLAREAV
MMAR_4928|M.marinum_M MLTAAAQLEGPVNDLAFSADTAESYSWKQDLGAAAPMGIPTRRTVVREAV
MUL_0477|M.ulcerans_Agy99 MLTAAAQLEGPVNDLAFSADTAESYSWKQDLGAAAPMGIPTRRTVVREAV
MAV_0713|M.avium_104 MLTSAAQLEGPVGDLSFAADTAESYEWNQDLGQASPMGIPTRRTIAREAV
Mflv_1600|M.gilvum_PYR-GCK MLTAAAQLEGPVDDLSFCADTAESYRWSTDLGIASPMGIKTRRTVTREAI
Mvan_5157|M.vanbaalenii_PYR-1 MLTSAAQLEGPVEDLSFCADTAESYAWTTDLGIASPMGIKTHRTIAREAV
MSMEG_5859|M.smegmatis_MC2_155 MLTSGAQLEGPIDDLAFSADTAENYPWRTDLGHATPQGIPTDRVIAREAV
TH_2814|M.thermoresistible__bu MLTSGGQLETPVEDLSFAADTAENYRWSTDLGRATPQGIPTDRRIVREPV
MAB_1218|M.abscessus_ATCC_1997 MLTAGAQLEGPVDDLQFAADTAESFDWTSDLGVASPMGIKTRRTLAREAV
MLBr_02573|M.leprae_Br4923 LQSAKDEAMKCAKGFRYYAEKAETLLADEPAEAAAVG--ASKALARYQPL
: :: : .: : *:.**. *: : :.:
Rv0768|M.tuberculosis_H37Rv GVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTPWCAAALGEIIV
Mb0791|M.bovis_AF2122/97 GVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTPWCAAALGEIIV
MMAR_4928|M.marinum_M GVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTPWCAAVLGELIA
MUL_0477|M.ulcerans_Agy99 GVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTPWCAAVLGELIA
MAV_0713|M.avium_104 GVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTPWCAAVLGQLIA
Mflv_1600|M.gilvum_PYR-GCK GVVGAITPWNFPHQINLAKVGPALAAGNTLVLKPAPDTPWAAAVLGELIT
Mvan_5157|M.vanbaalenii_PYR-1 GVVGAITPWNFPHQINLAKIGPALAAGNTLVLKPAPDTPWAAAVLGELIA
MSMEG_5859|M.smegmatis_MC2_155 GVVGAITPWNFPHQINLAKVGPALAAGNTLVLKPAPDTPWAAAVLGQIIT
TH_2814|M.thermoresistible__bu GVVGAITPWNFPHQINLAKIGPALAAGNTLVLKPAPDTPWAGAVLGELIT
MAB_1218|M.abscessus_ATCC_1997 GVVGAVTPWNFPHQINLAKVGPALAAGNTLVLKPAPDTPWCAAVLGRIIA
MLBr_02573|M.leprae_Br4923 GVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQCALYLADVIA
*** *: ***** .: .*** ***. :** *.:.* .. *. :*.
Rv0768|M.tuberculosis_H37Rv EHTDFPPGVVNIVTSSSHALGALLAKDPRVDMISFTGSTATGRAVMADAA
Mb0791|M.bovis_AF2122/97 EHTDFPPGVVNIVTSSSHALGALLAKDPRVDMISFTGSTATGRAVMADAA
MMAR_4928|M.marinum_M DCTDIPPGVVNIVTSSEHGLGALLAKDPRVDMVSFTGSTATGRSVMADGA
MUL_0477|M.ulcerans_Agy99 DCTDIPPGVVNIVTSSEHGLGALLAKDPRVDMVSFTGSTATGRSVMADGA
MAV_0713|M.avium_104 EHTDFPPGVVNIITSDDHGVGALLSKDPRVDMVSFTGSTVTGRSVMSDAA
Mflv_1600|M.gilvum_PYR-GCK EHTDFPAGVINIVTSSDHGVGALLSKDPRVDMVSFTGSTNTGRAVMADGA
Mvan_5157|M.vanbaalenii_PYR-1 EHTDFPAGVINIVTSSDHGVGALLSKDPRVDMVSFTGSTNTGRAVMADGA
MSMEG_5859|M.smegmatis_MC2_155 EHTDFPPGVINIVTSSDHSVGALLSKDPRVDMVSFTGSTATGRAVMTDAA
TH_2814|M.thermoresistible__bu GHTDFPPGVINIVTSSDHAVGAVLASDPRVDMVSFTGSTATGRSVMTDAA
MAB_1218|M.abscessus_ATCC_1997 EHTDFPPGVVNIVTSSDHRIGAQLSTDPRVDMVSFTGSTATGRAVMTDAA
MLBr_02573|M.leprae_Br4923 RG-GFPEDCFQTLLISANGVEAILR-DPRVAAATLTGSEPAGQAVGAIAG
.:* . .: : . : : * * **** ::*** :*::* : ..
Rv0768|M.tuberculosis_H37Rv ATIKKVFLELGGKSAFVVLDD---ADLAAASAVSAFSACMHAGQGCAITT
Mb0791|M.bovis_AF2122/97 ATIKKVFLELGGKSAFVVLDD---ADLAAASAVSAFSACMHAGQGCAITT
MMAR_4928|M.marinum_M ATIKRMFLELGGKSAFIVLDD---ADLAAASSVSAFTASMHAGQGCAITT
MUL_0477|M.ulcerans_Agy99 ATIKRMFLELGGKSAFIVLDD---ADLAAASSVSAFTASMHAGQGCAITT
MAV_0713|M.avium_104 ATIKRVFLELGGKSAFVVLDD---ADLGGACSMSAFTAAMHAGQGCAITT
Mflv_1600|M.gilvum_PYR-GCK ATLTKVFLELGGKSAFLVLDD---ADLAGACSMAAFTASMHAGQGCAITT
Mvan_5157|M.vanbaalenii_PYR-1 ATLKKVFLELGGKSAFLVIDD---ADLAGACSMAAFTASMHAGQGCAITT
MSMEG_5859|M.smegmatis_MC2_155 LTIKKVFLELGGKSAFLVLDD---ADLTGACSMAAFTVAMHAGQGCAITT
TH_2814|M.thermoresistible__bu ATIKKVFLELGGKSAFLVLDD---ADLAGACSMAAFTAAMHAGQGCAITT
MAB_1218|M.abscessus_ATCC_1997 ATVKKVFLELGGKSAFIVLDDLDEAALGSACSVAAFTAAMHAGQGCAITT
MLBr_02573|M.leprae_Br4923 NEIKPTVLELGGSDPFIVMPS---ADLDAAVATAVTGRVQNNGQSCIAAK
:. .*****...*:*: . * * .* : :. : **.* :.
Rv0768|M.tuberculosis_H37Rv RLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVCGPLISARQRDRVQGY
Mb0791|M.bovis_AF2122/97 RLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVCGPLISARQRDRVQGY
MMAR_4928|M.marinum_M RLVVPRARYDEAVAIAAGTMSSIKPGDPDDARTVCGPLISQRQRDRVQGY
MUL_0477|M.ulcerans_Agy99 RLVVPRARYDEAVAIAAGTMSSIKPGDPDDARTVCGPLISQRQRDRVEGY
MAV_0713|M.avium_104 RLLVPRDRYDEAVAAAAGTMGSIKPGDPNDPGTVCGPVISARQRDRVQGY
Mflv_1600|M.gilvum_PYR-GCK RLLVPRARYDEAVEAAAATMGGLKPGDPDKPGTICGPVISARQRDRIQSY
Mvan_5157|M.vanbaalenii_PYR-1 RLVVPRDRYDEAVEAAAATMGGLKAGDPTKPGTVCGPLISERQRERVQGY
MSMEG_5859|M.smegmatis_MC2_155 RLVVPRARYDEAVEIAAGTLGSIKPGDPTSKRTVCGPVISARQRDRVQSY
TH_2814|M.thermoresistible__bu RLVVPRARYDEAVEAAAATMGSLKPGDPTDKRTVCGPVISARQRDRVQGY
MAB_1218|M.abscessus_ATCC_1997 RLVVPRERYDDAVAAAAATMGSLKPGDPAKPGTICGPVISARQRDRVQSY
MLBr_02573|M.leprae_Br4923 RFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATESGRAQVEQQ
*::. *:: * .. : :: *** . * **: : * :::
Rv0768|M.tuberculosis_H37Rv LDLAVAEGGRFACGGARPADREVGFYIEPTVIAGLTNDARVAREEIFGPV
Mb0791|M.bovis_AF2122/97 LDLAVAEGGRFACGGARPADREVGFYIEPTVIAGLTNDARVAREEIFGPV
MMAR_4928|M.marinum_M LDLAIAEGGTFACGGGRPAGRQVGYFIEPTVIAGLTNDARPAREEIFGPV
MUL_0477|M.ulcerans_Agy99 LDLAIAEGGTFACGGGRPAGRQVGYFIEPTVIAGLTTDARPAREEIFGPV
MAV_0713|M.avium_104 LDLAIAEGGTFACGGGRPADKDVGFFIEPTVIAGLTNDARPAREEIFGPV
Mflv_1600|M.gilvum_PYR-GCK LDSAVAEGGRFACGGGRPADRDTGFFIEPTVIAGLDNNAKVAREEIFGPV
Mvan_5157|M.vanbaalenii_PYR-1 LDSAIAEGGRFACGGGRPADRDTGFFIDPTVIAGLDNNAKVAREEIFGPV
MSMEG_5859|M.smegmatis_MC2_155 LDLAIKEGGRFACGGGRPADMDKGFWIEPTVIAGLDNNARVAREEIFGPV
TH_2814|M.thermoresistible__bu LDLAVAEGGRFACGGGRPADRDVGFFIEPTVIAGLTNDARVAREEIFGPV
MAB_1218|M.abscessus_ATCC_1997 LDLAVAEGGSFACGGGRPEGKDRGFFIEPTVVAGLTNAARVAREEIFGPV
MLBr_02573|M.leprae_Br4923 VDAAAAAGAVIRCGGKRPD--RPGWFYPPTVITDITKDMALYTDEVFGPV
:* * *. : *** ** *:: ***::.: . :*:****
Rv0768|M.tuberculosis_H37Rv LTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRAARIASRLRVGTVNVN
Mb0791|M.bovis_AF2122/97 LTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRAARIASRLRVGTVNVN
MMAR_4928|M.marinum_M LTVLAHDGDDDAVRIANDSPYGLSGTVYGGDPQRAADVAARLRVGTVNVN
MUL_0477|M.ulcerans_Agy99 LTVLAHDGDDDAVRIANDSPYGLSGTVYGGDPQRAADVAARLRVGAVNVN
MAV_0713|M.avium_104 LTVIAYDGEEDALRIANDSPYGLSGTVFSADPERAANFASRMRVGTVNVN
Mflv_1600|M.gilvum_PYR-GCK LTVIAHDGDDDAVRIANDSPYGLSGTVFSPDLERANGVAARLRVGTVNIN
Mvan_5157|M.vanbaalenii_PYR-1 LTVIAHDGDDDAVRIANDSPYGLSGTVFSADPERAAGIAARMRVGTVNVN
MSMEG_5859|M.smegmatis_MC2_155 LTVIAHDGDDDAVRIANDSPYGLSGTVFSGDDARAQAVASRMRVGTVNVN
TH_2814|M.thermoresistible__bu LTVIAHDGDDDAVRIANDSPYGLSGTVFSGDDERAQAVAARLRVGTVNVN
MAB_1218|M.abscessus_ATCC_1997 LVVIAHDGDEDAVRIANDSPYGLSGTVFGDDDERSARVASRLRVGTVNVN
MLBr_02573|M.leprae_Br4923 ASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNIVAGQVFIN
* . . :**:.*** :*:**....: : . . .: .* * :*
Rv0768|M.tuberculosis_H37Rv GGVWYCADAPFGGYKQSGIGREMGLLGFEEYLEAKLIATAAN-
Mb0791|M.bovis_AF2122/97 GGVWYCADAPFGGYKQSGIGREMGLLGFEEYLEAKLIATAAN-
MMAR_4928|M.marinum_M GGVWYCADAPFGGYKQSGIGREMGLAGFEEYLETKLIATAAN-
MUL_0477|M.ulcerans_Agy99 GGVWYCADAPFGGYKQSGIGREMGLAGFEEYLETKLIATAAN-
MAV_0713|M.avium_104 GGVWYSADAPFGGYKQSGNGREMGLAGFEEYLEIKTIATAVQ-
Mflv_1600|M.gilvum_PYR-GCK GGVWYSADMPFGGYKQSGIGREMGLAGFEEYLEIKAIATAVN-
Mvan_5157|M.vanbaalenii_PYR-1 GGVWYSADMPFGGYKQSGIGREMGLAGFEEYLEIKAIATAAN-
MSMEG_5859|M.smegmatis_MC2_155 GGIWYSADAPFGGYKQSGVGREMGLAGFEEYTEIKLIATLAG-
TH_2814|M.thermoresistible__bu GGIWYSADAPFGGYKQSGIGREMGVAGFEEYTELKVIATAVAQ
MAB_1218|M.abscessus_ATCC_1997 GGVWYSADAPFGGYKQSGNGREMGVAGFEEYLETKVIATAVK-
MLBr_02573|M.leprae_Br4923 GMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTVWIA---
* .: **** *:** ***:. *:.*: : * :