For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GHLDADKKTSHQPIEGDCVSQGEATSVAVEGKGLKGGAVGLLSSVVIGTASTAPAYSLAATLGLIVASRG ELLCGVKAPAIVLLSFIPMYMIAVAYQELNKAEPDCGTTFTWASRAFGPVTGWLGGWGIIAADVIVMANL AQIAGAYSFTFVGELGWQSASNLAHSALWSTVAGVAWIVVMTYICYRGTDVSARMQYVLLSVELVVLVAL AAVALARVYTNHGDVGSLRPSLSWLWPSGLDFGTQMAPALLITIFIYWGFDTAVACNEETDDPGRTPGRA AVISTFLLLATYAVVTVASIAFAGVGVDGIGLGNPDNSNDVFAAVGPALFGDGVLGKIGLLLLSASILTS ASASTQTTILPTARTTLSMAVYKALPASFAKIHPRYLTPTVSTIAMGAVSILFYVLFTLVSRNLLSALIG SVGLVIAFYYGLTGYACAHYYRATLTKSLRNFVMRGVVPLTGGVVLTVVFIFGLIEYAKRDWLVENGHNV TIFGFGAVAVVGVGSMLLGVVVMLIWWAISPGFFRGRTLRRHSADLLLEPAVSPPAHLALPDSGDMQTVI AADLSNLPAGETAVNPQTGQHLTKDTSGAS*
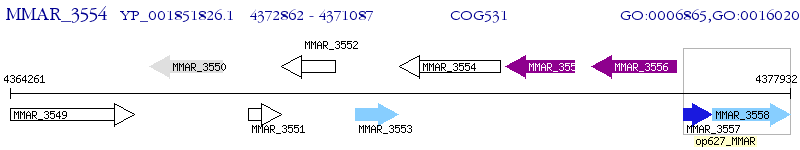
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3554 | - | - | 100% (591) | hypothetical protein MMAR_3554 |
| M. marinum M | MMAR_2609 | - | 1e-98 | 38.15% (540) | hypothetical protein MMAR_2609 |
| M. marinum M | MMAR_2243 | - | 1e-14 | 22.99% (435) | cationic amino acid transport integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2022c | - | 1e-26 | 27.44% (441) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_4853 | - | 1e-119 | 45.53% (492) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv1999c | - | 1e-26 | 27.44% (441) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3331c | - | 0.0 | 73.40% (564) | putative amino acid permease |
| M. avium 104 | MAV_2087 | - | 4e-12 | 23.43% (367) | amino acid transporter |
| M. smegmatis MC2 155 | MSMEG_1625 | - | 1e-107 | 41.18% (510) | amino acid transporter, putative |
| M. thermoresistible (build 8) | TH_4382 | - | 2e-10 | 23.27% (404) | PUTATIVE amino acid permease-associated region |
| M. ulcerans Agy99 | MUL_2787 | - | 0.0 | 99.15% (591) | hypothetical protein MUL_2787 |
| M. vanbaalenii PYR-1 | Mvan_1574 | - | 1e-124 | 46.03% (504) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2022c|M.bovis_AF2122/97 ----------------------MRRPLDPRDIPDELRRRLGLLDAVVIGL
Rv1999c|M.tuberculosis_H37Rv ----------------------MRRPLDPRDIPDELRRRLGLLDAVVIGL
MMAR_3554|M.marinum_M MGHLDADKKTSHQPIEGDCVSQGEATSVAVEGKGLKGGAVGLLSSVVIGT
MUL_2787|M.ulcerans_Agy99 MGHLDADKKTNHQPIEGDCVSQGEATSVAVEGKGLKGGAVGLLSSVVIGT
MAB_3331c|M.abscessus_ATCC_199 -----------------------MSSDIPVESKGLKGGALGLLSSVVVGL
Mflv_4853|M.gilvum_PYR-GCK ---------------------------------------------MVIGL
Mvan_1574|M.vanbaalenii_PYR-1 ---MAITDEPAVP---------------AVGGKGLQAGALGLVGNVVIGL
MSMEG_1625|M.smegmatis_MC2_155 ---MTRSDPQSDPESDPHHALEIVEQSAGLRGKGLAAGKVGTFSGAILGI
TH_4382|M.thermoresistible__bu -----------------------MSGTRTEQAQPELRRVLGPGLLLLFIV
MAV_2087|M.avium_104 ---------------------------MAPGTADHLRRGIGTFQLTMFGV
:.
Mb2022c|M.bovis_AF2122/97 GSMIGAGIFAALAPAAYAA------GSGLLLGLAVAAVVAYCNAISSARL
Rv1999c|M.tuberculosis_H37Rv GSMIGAGIFAALAPAAYAA------GSGLLLGLAVAAVVAYCNAISSARL
MMAR_3554|M.marinum_M ASTAPAYSLAATLGLIVASRGELLCGVKAPAIVLLSFIPMYMIAVAYQEL
MUL_2787|M.ulcerans_Agy99 ASTAPAYSLAATLGLIVASRGELLCGVKAPAIVLLSFIPMYMIAVAYQEL
MAB_3331c|M.abscessus_ATCC_199 ASTAPAYSLAATLGLIVASGGALLAGVKAPAIVLISFIPMYLIAVAYQEL
Mflv_4853|M.gilvum_PYR-GCK GAVAPAYSLAATLGYVVLN-----VQEKAPSMFVLAFIPMLLVAFAYKEL
Mvan_1574|M.vanbaalenii_PYR-1 GAVAPAYSLAATLGYVVLN-----VQEKAPAMFVLAFIPMLLVAFAYKEL
MSMEG_1625|M.smegmatis_MC2_155 SSVAPGYTLTASIGLIVAA-----VGLKMPAIFIAGFIPMFLTAYAYREL
TH_4382|M.thermoresistible__bu GDILGTGIYALVGDVAGEV------GGAAWLPFLIAFLIATVTAFSYLEL
MAV_2087|M.avium_104 GSTIGTGIFFVMSQAVPEA------GPAVIVSFLLAGVAAGLAAVCYAEL
. . . : * . .*
Mb2022c|M.bovis_AF2122/97 AARYPASGGTYVYGRMRLG-DFWGYLAGWGFVVGKTASCAAMALTVGFYV
Rv1999c|M.tuberculosis_H37Rv AARYPASGGTYVYGRMRLG-DFWGYLAGWGFVVGKTASCAAMALTVGFYV
MMAR_3554|M.marinum_M NKAEPDCGTTFTWASRAFG-PVTGWLGGWGIIAADVIVMANLAQIAGAYS
MUL_2787|M.ulcerans_Agy99 NKAEPDCGTTFTWASRAFG-PVTGWLGGWGIIAADVIVMANLAQIAGAYS
MAB_3331c|M.abscessus_ATCC_199 NKAEPDCGTTFTWASRTFG-PIVGWLGGWGIIAADVICLANLAQVAGAYS
Mflv_4853|M.gilvum_PYR-GCK SQDTPDCGTTFTWATKAFG-PWIGWIGGWGLAVSAIIVLANVSEIAAIYL
Mvan_1574|M.vanbaalenii_PYR-1 SQDTPDCGTTFTWGTKAFG-PWIGWIGGWGLAVSAIIVLANVSEIAAIYL
MSMEG_1625|M.smegmatis_MC2_155 NSRAPDCGASFTWSTKAFG-PYVGWMCGWGMVIATIIVLSNLAAIAVEFF
TH_4382|M.thermoresistible__bu VTKYPQAAGAALYVHKAFGIQFVTFLVAFIVMCSGITSASTASRFFAANL
MAV_2087|M.avium_104 ASAVPVSGSSYSYAYTTLG-EVVAMGVAACLLLEYGVATAAVSVNWSGYL
* .. : : :* . . : :
Mb2022c|M.bovis_AF2122/97 WPAQA-----------------------HAVAVAVVVALTAVNYAGIQKS
Rv1999c|M.tuberculosis_H37Rv WPAQA-----------------------HAVAVAVVVALTAVNYAGIQKS
MMAR_3554|M.marinum_M FTFVG-EL-----GWQSASNLAHSALWSTVAGVAWIVVMTYICYRGTDVS
MUL_2787|M.ulcerans_Agy99 FTFVG-EL-----GWQSASNLAHSTLWSTVAGVAWIVVMTCICYRGTDVS
MAB_3331c|M.abscessus_ATCC_199 FTFVG-DL-----GWSAASGLADSVLWSTVAGVIWIVIMTYICYRGIEVS
Mflv_4853|M.gilvum_PYR-GCK YNFLG------------LTSLAESVPATVALGCFFIIAMTLVSARGIVVS
Mvan_1574|M.vanbaalenii_PYR-1 YNFLG------------LDSLAESVPATVALGCFFIISMTLVSARGIVVS
MSMEG_1625|M.smegmatis_MC2_155 YLFVARIL-----NTPEIADLADNKLVNILTTLVFIALATWVSSRGITTS
TH_4382|M.thermoresistible__bu EVGLG---------------QEWGRLGVAGVALLFMALLAMVNLRGVGES
MAV_2087|M.avium_104 NKLLSNVVGFQLPHALSAAPWDAQPGYVNLPAVMLIGMCALLLIRGASES
. : : : * *
Mb2022c|M.bovis_AF2122/97 AWLTRSIVAVVLVVLTAVVVAAYG---SGAADPARLDIGVDAHVWG----
Rv1999c|M.tuberculosis_H37Rv AWLTRSIVAVVLVVLTAVVVAAYG---SGAADPARLDIGVDAHVWG----
MMAR_3554|M.marinum_M ARMQYVLLSVELVVLVALAAVALARVYTNHGDVGSLRPSLSWLWP-SGLD
MUL_2787|M.ulcerans_Agy99 ARMQYVLLSVELVVLVALAAVALARVYTNHGDVGSLRPSLSWLWP-SGLD
MAB_3331c|M.abscessus_ATCC_199 ARLQYWLLTVEVVVLIAFAVVALIKVYTHNAESYSLLPSLSWFWP-GGMD
Mflv_4853|M.gilvum_PYR-GCK ERVQNVLIAIQFGVLITVSIIALVRVFSGTAGAQAVMPEVSWLWP-SGLD
Mvan_1574|M.vanbaalenii_PYR-1 ERVQNVLIAIQFGVLITVSVIALIRVFTGTAGAQAISPQLSWLWP-SGLD
MSMEG_1625|M.smegmatis_MC2_155 EHVQYVLVGFQMTVLLAFAITAVIHVGAGNA-PEGLAFDLDWFNPFTNLA
TH_4382|M.thermoresistible__bu VRLNVLLTIVEISGLLLVIFVGLWAFTGGGADVDFTR--VVAFETTTDKS
MAV_2087|M.avium_104 AKVNAIMVMVKLGVLVVFGILAFTAFDMHHLDDFAPFG------------
: : . . * . .
Mb2022c|M.bovis_AF2122/97 ----MLQAAGLLFFAFAGYARIATLGEEVRDPARTIPRAIPLALGITLAV
Rv1999c|M.tuberculosis_H37Rv ----MLQAAGLLFFAFAGYARIATLGEEVRDPARTIPRAIPLALGITLAV
MMAR_3554|M.marinum_M FGTQMAPALLITIFIYWGFDTAVACNEETDDPGRTPGRAAVISTFLLLAT
MUL_2787|M.ulcerans_Agy99 FGTQMAPALLITIFIYWGFDTAVACNDETDDPGRTLGRAAVISTFLLLAT
MAB_3331c|M.abscessus_ATCC_199 FGTVIAPAVLTTIFIYWGWDTAVACNEESDDPGRTPGRAAVISTFLLLAT
Mflv_4853|M.gilvum_PYR-GCK VS-SIAAAVILCIFIYWGWDACLAVGEETKDPGKTPGIAALITTVILVCT
Mvan_1574|M.vanbaalenii_PYR-1 VS-SIAAAVILCIFIYWGWDACLAVGEETKDPGRTPGIAALVTTVILVCT
MSMEG_1625|M.smegmatis_MC2_155 FG-AFVIGVTGSIFAFWGWDTCLTLGEESKDPTKVPGRAGLLCVTTILLT
TH_4382|M.thermoresistible__bu VFLAVSAATSLAFFAMVGFEDSVNMAEETRDPVVIFPRALLTGLGIAGAV
MAV_2087|M.avium_104 -VAGVGTAAGTIFFSYIGLDAVSTAGDEVTNPQKTMPRALIAALLTVTGV
. . :* * :* :* * .
Mb2022c|M.bovis_AF2122/97 YALVAVAVIAVLGPQRLARAAAPLSEAMRVA---G---------VNWLIP
Rv1999c|M.tuberculosis_H37Rv YALVAVAVIAVLGPQRLARAAAPLSEAMRVA---G---------VNWLIP
MMAR_3554|M.marinum_M YAVVTVASIAFAGVGVDGIGLGNPDNSNDVFAAVGPALFGDGVLGKIGLL
MUL_2787|M.ulcerans_Agy99 YAVVTVASIAFAGVGVDGIGLGNPDNSNDVFAAVGPALFGDGVLGKIGLL
MAB_3331c|M.abscessus_ATCC_199 YALVSVAAVAFAGVGTEGVGLGNQENAADVFKAIGPELFGDSFVGHLGML
Mflv_4853|M.gilvum_PYR-GCK YVLVAYALQSFAGFSEVGIGLNNPNNTDDVLTVLG-----EPVAGAIAAS
Mvan_1574|M.vanbaalenii_PYR-1 YVLVAFALQSFAGFSEVGIGLNNPNNTDDVLTVLG-----KPVAGAIAAS
MSMEG_1625|M.smegmatis_MC2_155 YLLVAVAVMMYAGVGETGLGLGNPENSENVFGALA-----DPVLGSWGGP
TH_4382|M.thermoresistible__bu YVVVAIVAVALVPVGTLQASDVPLVEVVKAG-----------APGLPIEE
MAV_2087|M.avium_104 YVFVALAALGTQPWQDFGGQQEAGLATILDHVTHG----------SWAST
* .*: .
Mb2022c|M.bovis_AF2122/97 VVQIGAAVAALGSLLALILGVSRTTLAMARDRHLPRWLAAVHPRFKVPFR
Rv1999c|M.tuberculosis_H37Rv VVQIGAAVAALGSLLALILGVSRTTLAMARDRHLPRWLAAVHPRFKVPFR
MMAR_3554|M.marinum_M LLSASILTSASASTQTTILPTARTTLSMAVYKALPASFAKIHPRYLTPTV
MUL_2787|M.ulcerans_Agy99 LLSASILTSASASTQTTILPTARTTLSMAVYKALPASFAKIHPRYLTPTV
MAB_3331c|M.abscessus_ATCC_199 LLSASILTSASACTQTTILPTARTTLSMGVYKALPDSFASIHRTYLTPST
Mflv_4853|M.gilvum_PYR-GCK ALLLTVSVSALSSTQTTILPTARGTLSMAVYEAIPRRFASVHPRYMTPAF
Mvan_1574|M.vanbaalenii_PYR-1 ALLLTVSVSALSSTQTTILPTARGTLSMAVYEAIPKRFANVHPRYMTPAF
MSMEG_1625|M.smegmatis_MC2_155 LLFLAVLASSVASLQTTFLPAARAMLAMGAYGAFPKRFSHVHPRFLVPSF
TH_4382|M.thermoresistible__bu LLPWISMFAVSNTALINMLMASRLIYGMARQRVLPPVLGSVHPRRRTPWV
MAV_2087|M.avium_104 ILAAGAVISIFSVTLVTMYGITRILFAMGRDGLLPPRFARVNPRTMTPVN
: : : :* .*. :* :. :: .*
Mb2022c|M.bovis_AF2122/97 AELVVG------AVVAALAATADIRGAIG-FSSFGVLVYYAIAN------
Rv1999c|M.tuberculosis_H37Rv AELVVG------AVVAALAATADIRGAIG-FSSFGVLVYYAIAN------
MMAR_3554|M.marinum_M STIAMGAVSILFYVLFTLVSRNLLSALIG-SVGLVIAFYYGLTGYACAHY
MUL_2787|M.ulcerans_Agy99 STIAMGAVSILFYVLFTLVSRNLLSALIG-SVGLVIAFYYGLTGYACAHY
MAB_3331c|M.abscessus_ATCC_199 STIAMGAISIVFYVLFTAISTNLLTALIG-SVGLMIAFYYGLTGFACAWF
Mflv_4853|M.gilvum_PYR-GCK GTIVMGISALFFYLVLKMLSENALLDSVA-SLGLAVAFYYGITAFACVWY
Mvan_1574|M.vanbaalenii_PYR-1 GTIVMGLSALFFYLVLKALSENALLDSVA-SLGLAVAFYYGITAFACVWY
MSMEG_1625|M.smegmatis_MC2_155 STLIAGIVTGVFYTVVSLLSEYALLDTIA-ALGIMICWYYGITAFACIWY
TH_4382|M.thermoresistible__bu AIIFTTLIAFGLIFYVTAFADSTAIAVLGGTTSLLLLAVFAMVNVAVLVL
MAV_2087|M.avium_104 NTVIVA-------VAAATLAAFIPLQNLADMVSIGTLTAFVVVSVGVIVL
: : :. .: : :.
Mb2022c|M.bovis_AF2122/97 -ASALTLGLDEGRPRRLIPLVG-LIGCVVLAFALPLSSVA----------
Rv1999c|M.tuberculosis_H37Rv -ASALTLGLDEGRPRRLIPLVG-LIGCVVLAFALPLSSVA----------
MMAR_3554|M.marinum_M YRATLTKSLRNFVMRGVVPLTGGVVLTVVFIFGLIEYAKRDWLVEN-GHN
MUL_2787|M.ulcerans_Agy99 YRATLTKSLRNFVMRGVVPLTGGVVLTVVFIFGLIEYAKRDWLVEN-GHN
MAB_3331c|M.abscessus_ATCC_199 YRKRLTTSFRDLMMRGVMPLAGGATLTVVFIYGLIQYAKPNWLIDDDGNN
Mflv_4853|M.gilvum_PYR-GCK FRRTLFSSARNLFFRGVFPLLGGLSMLAAFAISAKDMIQP-----DYG-H
Mvan_1574|M.vanbaalenii_PYR-1 FRRTLFESTRNLFFRGVFPLLGGLSMLAAFTISAKDMIQP-----DYG-Y
MSMEG_1625|M.smegmatis_MC2_155 FRRELFTDAHNIVFKFLFPLIGGLVLAAVFGFSLKESMDP-----ENGSG
TH_4382|M.thermoresistible__bu RRDESRDPVVARRHFRTPTFLPVVGFVASLYLVTPLSGRP----------
MAV_2087|M.avium_104 RVREP--DLPRGFRVPGYPVTPVLSIMACGYILASLHWYT----------
.. .
Mb2022c|M.bovis_AF2122/97 ------AGAAVLGVGVAAYGVRRIITRRAR---QTDSGDTQRSGHPSAT-
Rv1999c|M.tuberculosis_H37Rv ------AGAAVLGVGVAAYGVRRIITRRAR---QTDSGDTQRSGHPSAT-
MMAR_3554|M.marinum_M VTIFGFGAVAVVGVGSMLLGVVVMLIWWAI-SPGFFRGRTLRRHSADLLL
MUL_2787|M.ulcerans_Agy99 VTIFGFGAVAVVGVGSMLLGVVVMLIWWAI-SPGFFRGRTLRRHSADLLL
MAB_3331c|M.abscessus_ATCC_199 VTVFGVGAVAVVGVGALVLGVVLMMLWRVA-APPFFQGQTLPRRSADLVL
Mflv_4853|M.gilvum_PYR-GCK TAFGSIGGVFVLGVGMLVLGIPLMLACFAFGTKRFFRGETLNADTAVKVP
Mvan_1574|M.vanbaalenii_PYR-1 TAFGPVGGVFVLGVGMLVLGIPLMLACFAFGTKRFFRGETLNADTEVKVP
MSMEG_1625|M.smegmatis_MC2_155 AELGGIGLVFYIGFGILLLGAVLMLVMRTR-SPEFFRGETLSRNTPVIPG
TH_4382|M.thermoresistible__bu -AQQYLLSVVLIVIGVLLFGVTMAINRQLG-IPTRAVGDPAKMVLPPD--
MAV_2087|M.avium_104 ---------WIAFSGWVLLALIFYFVWGRH---HSALNDAAVDPSGQER-
* . : . .
Mb2022c|M.bovis_AF2122/97 --------------------------------------------------
Rv1999c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3554|M.marinum_M EPAVSPPAHLALPDSGD-MQTVIAADLSNLPAGETAVNPQTGQHLTKDTS
MUL_2787|M.ulcerans_Agy99 EPAVSPPAHLALPDSGD-MQTVIAADLSNLPAGETAVNPQTGQHLTKDTS
MAB_3331c|M.abscessus_ATCC_199 APAAAT-THFGLPDAVEAQQTVIAGDLSNLPEGEVAVNPQTGEEFTKD--
Mflv_4853|M.gilvum_PYR-GCK DTF-----------------------------------------------
Mvan_1574|M.vanbaalenii_PYR-1 DTF-----------------------------------------------
MSMEG_1625|M.smegmatis_MC2_155 --------------------------------------------------
TH_4382|M.thermoresistible__bu --------------------------------------------------
MAV_2087|M.avium_104 --------------------------------------------------
Mb2022c|M.bovis_AF2122/97 ---
Rv1999c|M.tuberculosis_H37Rv ---
MMAR_3554|M.marinum_M GAS
MUL_2787|M.ulcerans_Agy99 GAS
MAB_3331c|M.abscessus_ATCC_199 ---
Mflv_4853|M.gilvum_PYR-GCK ---
Mvan_1574|M.vanbaalenii_PYR-1 ---
MSMEG_1625|M.smegmatis_MC2_155 ---
TH_4382|M.thermoresistible__bu ---
MAV_2087|M.avium_104 ---