For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SQPTGEPRLRRALGVAGLVAFGLAYIDPLAMFDTFGVVSQTTHGHVPTAYVVTFVAMLFTAASYAVLVHA YPSAGSAYTFARRSFGGNVGFLTGWALMLDYLLLPLVNYLLIGIYLNAQFPAVPAPVFCLAAIALVTTVN IVGVVIMNRLNFALVGLQLAFVLVFITVSLLYLRDHPDASLLQPLYSAGFRGGDILGGAAVVALSFVGFD AVSTMAEEARNPRRTVPLAIILTVLIGGATFLSVSYCATLVEPDYRAITTPNSAALQIVVSAGGHSLQTF FIIAYMCACSAAALSSQVSVTRILYAMGRDGVLPRAVFGRISRRFRTPIGAALVVSAVSTLALIADLETM ASIVSFGALAAFTLVHLAVTKHFLIDQRRRGWRSWALYAAMPAVGVVLTGWLWTSLSVGAFAIGFAWLGI GTSYLAVLTRGFRKPAPAVDFDADAAPIQTEIDLTKEGTLQ*
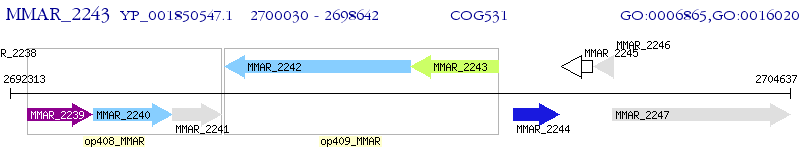
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2243 | - | - | 100% (462) | cationic amino acid transport integral membrane protein |
| M. marinum M | MMAR_0869 | - | 1e-85 | 39.16% (429) | amino acid permease |
| M. marinum M | MMAR_3621 | rocE | 7e-30 | 27.54% (443) | cationic amino acid transport integral membrane protein RocE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3281c | - | 8e-30 | 24.67% (458) | cationic amino acid transport integral membrane protein |
| M. gilvum PYR-GCK | Mflv_4427 | - | 8e-32 | 26.97% (456) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv3253c | - | 8e-30 | 24.67% (458) | cationic amino acid transport integral membrane protein |
| M. leprae Br4923 | MLBr_01305 | ansP2 | 7e-15 | 21.28% (470) | putative L-asparagine permease |
| M. abscessus ATCC 19977 | MAB_3599c | - | 1e-30 | 26.75% (456) | putative amino acid permease |
| M. avium 104 | MAV_4216 | - | 6e-32 | 27.31% (465) | cationic amino acid transporter |
| M. smegmatis MC2 155 | MSMEG_0446 | - | 1e-119 | 48.74% (437) | putrescine importer |
| M. thermoresistible (build 8) | TH_0471 | - | 1e-28 | 25.77% (454) | POSSIBLE CATIONIC AMINO ACID TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_2597 | - | 2e-29 | 25.93% (459) | cationic amino acid transport integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_1740 | - | 1e-32 | 26.30% (460) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2243|M.marinum_M ------------------MS--QPTGEP--RLRRALGVAGLVAFGLAYID
MSMEG_0446|M.smegmatis_MC2_155 ------------------MSSTAEVAEG--HLKRALGLPSLVLFGLVYMV
TH_0471|M.thermoresistible__bu --------MTDRLRVKSVEQSIADTDEPDSRLRKDLSWWDLTVFGVSVVI
Mvan_1740|M.vanbaalenii_PYR-1 MVQEPGAQTVGRRRTKSVEQSIADTDEPSTRLRKELNWWDLTVFGVSVVI
MAB_3599c|M.abscessus_ATCC_199 ------MPGSGLWRTKSVEQSIADTDEPETKLRRDLTWWDLTVFGVSVVI
Mb3281c|M.bovis_AF2122/97 --------MAGRRRMKSVEQSIADTDEPTTRLRKDLTWWDLVVFGVSVVI
Rv3253c|M.tuberculosis_H37Rv --------MAGRRRMKSVEQSIADTDEPTTRLRKDLTWWDLVVFGVSVVI
MUL_2597|M.ulcerans_Agy99 --------MAGGLRTKSVEQSIADTDEPETRLRKDLTWRDLVVFGVSVVI
MAV_4216|M.avium_104 --------MASRWRTKSVEQSIADTDEPDTRLRKDLTWWDLVVFGVAVVI
Mflv_4427|M.gilvum_PYR-GCK -----------------MSVDTAEGTAEPTRLARRITGPLLFLFILGDVL
MLBr_01305|M.leprae_Br4923 ------MATLAESPEPKSGASRAGVLGEEAGYHKGLKPRQLQMIGIGGAI
: : * : :
MMAR_2243|M.marinum_M PLAMFDTFG-VVSQTTHGHVPTAYVVTFVAMLFTAASYAVLVHAYPSAGS
MSMEG_0446|M.smegmatis_MC2_155 PLTVFTTYG-IVTQTSGGRVPLSYLVTLAAMVFTARSYARMSVAYPVAGS
TH_0471|M.thermoresistible__bu GAGIFTVTASTAGNITGPAISVSFIIAAIACGLAALCYAEFASTVPVAGS
Mvan_1740|M.vanbaalenii_PYR-1 GAGIFTITASTAANITGPAISLSFIIAAIACGLAALCYAEFASTVPVAGS
MAB_3599c|M.abscessus_ATCC_199 GAGIFTVTASTAANVTGPAISLSFIMAAIGCGLAAMCYAEFASTIPVAGS
Mb3281c|M.bovis_AF2122/97 GAGIFTVTASTAGDITGPAIWISFLIAAATCALAALCYAEFASTLPVAGS
Rv3253c|M.tuberculosis_H37Rv GAGIFTVTASTAGDITGPAIWISFLIAAATCALAALCYAEFASTLPVAGS
MUL_2597|M.ulcerans_Agy99 GAGIFTVTASTAGDITGPAIWISFLIAAGTCALAALCYAEFASTLPVAGS
MAV_4216|M.avium_104 GAGIFTVTASTTGDITGPAIWVSFVIAAGTCALAALCYAEFASTLPVAGS
Mflv_4427|M.gilvum_PYR-GCK GAGIYALMGELSSEVGG-VLWAPMTVALGLALLTAGSYAELVTKYPRAGG
MLBr_01305|M.leprae_Br4923 GTGLFLGAG-GRLAKAGPGLFLVYAVCGVFVFLILRALGELVLHRPSSGS
:: . : : : . . : * :*.
MMAR_2243|M.marinum_M AYTFARRSFG-GNVGFLTGWALMLDYLLLPLVNYLLIG-IYLN-------
MSMEG_0446|M.smegmatis_MC2_155 AYTYTQKSFG-APVGFLAGWSLLLDYLFLPMINYLVIG-LYLS-------
TH_0471|M.thermoresistible__bu AYTFSYATFG-EFVAWIIGWDLILEFSVAAAVVAKGWS-SYLGEVFEFAG
Mvan_1740|M.vanbaalenii_PYR-1 AYTFSYATFG-EFVAWIIGWDLILEFAVAAAVVAKGWS-SYLGTVFGFAG
MAB_3599c|M.abscessus_ATCC_199 AYTFSYATFG-EFAAWILGWDLILEFSVGAATVAKGWS-SYLSTVFGLSS
Mb3281c|M.bovis_AF2122/97 AYTFSYATFG-EFLAWVIGWNLVLELAMGAAVVAKGWS-SYLGTVFGFGN
Rv3253c|M.tuberculosis_H37Rv AYTFSYATFG-EFLAWVIGWNLVLELAMGAAVVAKGWS-SYLGTVFGFGN
MUL_2597|M.ulcerans_Agy99 AYTFSYATFG-EFLAWVIGWNLVLELAIGAAVVAKGWS-SYLGTVFGFSG
MAV_4216|M.avium_104 AYTFSYATFG-EFLAWIIGWNLLLELAIGAAVVAKGWS-SYLGTVFGFSG
Mflv_4427|M.gilvum_PYR-GCK AAVFAERAFGKPLISFLVGFSMLAAGVTSAAGLALAFSGDYLAT------
MLBr_01305|M.leprae_Br4923 FVSYAREFFG-EKAAYVVGWLYFLDWAMTAIVDTTAIA-TYLHR------
:: ** .:: *: . . . **
MMAR_2243|M.marinum_M --AQFPAVPAPVFCLAAIALVTTVNIVGVVIMNRLNFALVGLQLAFVLVF
MSMEG_0446|M.smegmatis_MC2_155 --AAIPAIPAWVFILIAIAIVTVLNIVGIVSVARANFVIIAVQAIFIVAF
TH_0471|M.thermoresistible__bu ATTRLGPITLDWGALLIIGFVTVLLATGTKLSANVSLAITVIKVGVVLLV
Mvan_1740|M.vanbaalenii_PYR-1 GTTEVAGFQLDWGALVIIALVTLILVMGTKLSAGVSLAITAVKVSVVLLV
MAB_3599c|M.abscessus_ATCC_199 GAVHLGAVKIDWGALLIVSVLTVLLAVGTKLSSRVSLVITTIKVLVVLLV
Mb3281c|M.bovis_AF2122/97 GTGHLGSLQLDWGALVIVTLVATLIALGTKLSSRFSAVVTAIKVSVVVLV
Rv3253c|M.tuberculosis_H37Rv GTGHLGSLQLDWGALVIVTLVATLIALGTKLSSRFSAVVTAIKVSVVVLV
MUL_2597|M.ulcerans_Agy99 GTADFGSIRLDWGALLSVSIVSLLVAYGTKLSSRFSAVVTTIKVAVVILV
MAV_4216|M.avium_104 GTVKFGAAQLDWGALVIVGGVATLIALGTKLSSRFSAVITGIKVSVVVFV
Mflv_4427|M.gilvum_PYR-GCK ----FVDVPVALAAVVFLALVACLNARGISESLKTNVVMTAIELSGLVLV
MLBr_01305|M.leprae_Br4923 -WTIFTALPQWTLALLALAVVLVMNLISVEWFGELEFWAALIKVCALMAF
. : : : : . . :: :: .
MMAR_2243|M.marinum_M ITVSLLYLRDHPDASLLQP-------------------LYSAGFRGGD--
MSMEG_0446|M.smegmatis_MC2_155 LILATSKVFSYGTVDLAAP-------------------FTGDGTAPGFSP
TH_0471|M.thermoresistible__bu VIVGAFFIKAANYTPFIPPAEAGGEAGRSLDQSLFSLITGAEGSSYGWYG
Mvan_1740|M.vanbaalenii_PYR-1 VIVGAFYIKAENFSPFVPAPEEGGSGGTGTEQSLFSLLTGAEGSHYGWYG
MAB_3599c|M.abscessus_ATCC_199 IIVGAFFIKTANYTPFIPPAEAGSSSRQGLDQSLFSFVAGGSGSQYGWFG
Mb3281c|M.bovis_AF2122/97 VVVGAFYIRAANYSPFIPEPEVQHHGG-GLDQSVFSLLTGAQGSHYGWYG
Rv3253c|M.tuberculosis_H37Rv VVVGAFYIRAANYSPFIPEPEVQHHGG-GLDQSVFSLLTGAQGSHYGWYG
MUL_2597|M.ulcerans_Agy99 VVVGAFYVKAANYSPFIPEPEAGPEGR-GADQSLLSLLTGAPGSHYGWYG
MAV_4216|M.avium_104 VVVGVFYIKRANYSPFIPKPEAGGQAK-GIDQSVLSLLTGAHTSHYGWYG
Mflv_4427|M.gilvum_PYR-GCK IAVVAMMVGRGDGE--------------------VGRVTEFPDGTTPALA
MLBr_01305|M.leprae_Br4923 LVVGTIFLGGRYPVDGHNTG-------------LSLWTSHGGLFPTGVAP
: : :
MMAR_2243|M.marinum_M ILGGAAVVALSFVGFDAVSTMAEEARNPRRTVPLAIILTVLIGGATFLSV
MSMEG_0446|M.smegmatis_MC2_155 VLAGAAILCLSFLGFDAVSTLSEEAKDPKRTVPRAIMIATLASGLIFVVL
TH_0471|M.thermoresistible__bu LLAGASIVFFAFIGFDIIATTAEETKNPQRDVARGIMATLGIVTVLYVAV
Mvan_1740|M.vanbaalenii_PYR-1 VLAGASIVFFAFIGFDIVATTAEETRNPQRDVPRGILASLAIVTVLYVAV
MAB_3599c|M.abscessus_ATCC_199 VLAGASIVFFAFIGFDVVATTAEETRNPQKDVPRGIIASLVIVTVLYVAV
Mb3281c|M.bovis_AF2122/97 VLAGASIVFFAFIGFDIVATMAEETKRPQRDVPRGILASLGVVTLLYVAV
Rv3253c|M.tuberculosis_H37Rv VLAGASIVFFAFIGFDIVATMAEETKRPQRDVPRGILASLGVVTLLYVAV
MUL_2597|M.ulcerans_Agy99 VLAGASIVFFAFIGFDIVATMAEETKHPQRDVPRGILVSLGVVTLLYVAV
MAV_4216|M.avium_104 VLAGASIVFFAFIGFDIVATMAEETKHPQRDVPRGILASLGIVTLLYVAV
Mflv_4427|M.gilvum_PYR-GCK ILGGAILAYYSFVGFETSANVVEEIRDPRRVYPRALFGALITAGVVYALV
MLBr_01305|M.leprae_Br4923 LIVVSSGVMFAYAAVELVGTAAGETVEPKKIMPRAINSVIARIAIFYVGS
:: : :: ..: .. * *:: . .: :
MMAR_2243|M.marinum_M SYCAT--LVEPDYRAITTPNSAALQIVVSAGG-HSLQTFFIIAYMCACSA
MSMEG_0446|M.smegmatis_MC2_155 SYVAQ--VVFPSN-EFSDVDSGSLDVMMTAGG-NFLQVFFTAAYVAGATG
TH_0471|M.thermoresistible__bu SLVLTGMVHYTELRAAG--ENANLATAFAANGIDWAAKVISIGALAGLTT
Mvan_1740|M.vanbaalenii_PYR-1 AVVLTGMVSYTELRDA---ETQNLATAFSLNGVDWAAKVISIGALAGLTT
MAB_3599c|M.abscessus_ATCC_199 TVVLSGMVKYSDLRVAD--SHPNLSTAFHLNGVDWAAKVIAVGALAGLTT
Mb3281c|M.bovis_AF2122/97 SVVLSGMVPYTQLRTVPGRGPANLATAFQANGVYWASGIISVGALAGLTT
Rv3253c|M.tuberculosis_H37Rv SVVLSGMVPYTQLRTVPGRGPANLATAFQANGVYWASGIISVGALAGLTT
MUL_2597|M.ulcerans_Agy99 SIVLSGMVSYTQLRTVSGGGHANLATAFKANGVYWASGIISVGALAGLTT
MAV_4216|M.avium_104 AVVLSGMVSYTQLKTMPGRGQANLATAFTANGIHWASKIISIGALAGLTT
Mflv_4427|M.gilvum_PYR-GCK GLASAIAVPTDELSQSS----GPLLDVVAASGAAVPDWLFSLIALIAVAN
MLBr_01305|M.leprae_Br4923 VILLALLLPYSAFKASE----SPFVTFFSKVGFYGAGDLMNIVVLTAALS
: : . * .: : .
MMAR_2243|M.marinum_M AALSSQVSVTRILYAMGRDGVLPRAVFGRISRRFRTPIGAALVVSAVSTL
MSMEG_0446|M.smegmatis_MC2_155 SAITSQASVARILYAMGRDGVLPRPFFGHVSPRFSTPVYAILAVAVISLL
TH_0471|M.thermoresistible__bu VVIVLVLGQTRVLFAMSRDGLLPR-PLSKTGPRGTPVRITVLVGALVAIA
Mvan_1740|M.vanbaalenii_PYR-1 VVIVLVLGQTRVLFAMSRDGLLPR-QLAKTGEHGTPVRITLIVGAVVAVT
MAB_3599c|M.abscessus_ATCC_199 VVMVLMLGQIRVIFAMCRDGLLPR-ELARTGSHGTPIRITLIVGFLVAVA
Mb3281c|M.bovis_AF2122/97 VVMVLMLGQCRVLFAMARDGLVPR-QLAKTGSRGTPVRVTVLVAVLVATT
Rv3253c|M.tuberculosis_H37Rv VVMVLMLGQCRVLFAMARDGLVPR-QLAKTGSRGTPVRVTVLVAVLVATT
MUL_2597|M.ulcerans_Agy99 VVMVLMLGQCRVLFAMARDGLLPR-QLAKTGARGTPVRITVLVAVVVGAT
MAV_4216|M.avium_104 VVMVLMLGQCRVLFAMARDGLLPR-ALAKTGSRGTPVRITVLVALVVAVT
Mflv_4427|M.gilvum_PYR-GCK GALLTMIMASRLTFGMSDQGLLPSVFARVLPNRRTPWVAIVATTAVAMAL
MLBr_01305|M.leprae_Br4923 SLNAGLYATGRVMHSIAINGSGPK-FTARMSKNGVPYGGILLAAVICLCG
*: ..: :* * . . .
MMAR_2243|M.marinum_M ALIADLE------TMASIVSFGALAAFTLVHLAVTKHFLIDQRRRGWRSW
MSMEG_0446|M.smegmatis_MC2_155 AIVIDLG------ILASVISFGALVAFSAVNLSVIKHYFVDHRERAGAQA
TH_0471|M.thermoresistible__bu ASVFPVG------NLEEMVNIGTLFAFALVSAGVIVLRRTRPDLKRGFRV
Mvan_1740|M.vanbaalenii_PYR-1 ATVFPIG------KLEEMVNIGTLFAFVLVSAGVLVLRRTRPDLKRGFRT
MAB_3599c|M.abscessus_ATCC_199 ASLTPIE------GLEEIVNVGTLFAFVLVSAGVIVLRRSRPDLPRSFRT
Mb3281c|M.bovis_AF2122/97 ASVFPIT------KLEEMVNVGTLFAFILVSAGVVVLRRTRPDLQRGFTA
Rv3253c|M.tuberculosis_H37Rv ASVFPIT------KLEEMVNVGTLFAFILVSAGVVVLRRTRPDLQRGFTA
MUL_2597|M.ulcerans_Agy99 ASVFPIT------KLEEMVNVGTLFAFVLVSAGVIVLRKKRPDLERGFRA
MAV_4216|M.avium_104 ASVFPIT------KLEEMVNVGTLFAFVLVSAGVMVLRRTRPDLERGFKA
Mflv_4427|M.gilvum_PYR-GCK TLVGDLS------MLAETVVLLLLVVFLSANVSVLVLRRDHVEQD---HF
MLBr_01305|M.leprae_Br4923 VALNAFNPGQAFEIVLSVAALGIIAGWGTIVLCQLRLHKMAKAGIMRRPR
. : . : . . : :
MMAR_2243|M.marinum_M ALYAAMPAVGVVLTGWLWTSLSVGAFAIGFAWLGIG----------TSYL
MSMEG_0446|M.smegmatis_MC2_155 INSLVLPLIGFALTVWLWFSLDGLALIIGLCWLAIG----------FVWL
TH_0471|M.thermoresistible__bu PFVPWLPLAAIAACLWLMLNLTGLTWVRFLGWMALG----------LVLY
Mvan_1740|M.vanbaalenii_PYR-1 PWVPLLPILAIAACVWLMLNLTGLTWIRFLIWMAIG----------VVVY
MAB_3599c|M.abscessus_ATCC_199 PGVPWLPIVAIVACGWLMLNLAVRTWLFALSWMVIG----------VIVY
Mb3281c|M.bovis_AF2122/97 PWVPLLPIAAVCACLWLMLNLTALTWIRFGIWLVAG----------TAIY
Rv3253c|M.tuberculosis_H37Rv PWVPLLPIAAVCACLWLMLNLTALTWIRFGIWLVAG----------TAIY
MUL_2597|M.ulcerans_Agy99 PWVPVLPIASLCACLWLMLNLTALTWIRFGVWLLVG----------TVIY
MAV_4216|M.avium_104 PWVPVLPIASIGACVWLMLNLTALTWVRFGIWLVVG----------TAIY
Mflv_4427|M.gilvum_PYR-GCK RVWTFVPVLGIASCLLLLTQQTAKVWLLAAALLAVG----------VVLY
MLBr_01305|M.leprae_Br4923 FRMPLAPYSGYLTLAFLFAVLVVMAFDKPIGTWTVASLIVIVPALIAGWY
* . * . .
MMAR_2243|M.marinum_M AVLTRGFRKPAPAVDFDADAAPIQTEIDLTKEGTLQ---
MSMEG_0446|M.smegmatis_MC2_155 AGVTRGFQRPTPVLDMKE---------------------
TH_0471|M.thermoresistible__bu FFYGRHHSMLGRGMTDISARRTG----------------
Mvan_1740|M.vanbaalenii_PYR-1 FLYGRRHSLVGRRMG------------------------
MAB_3599c|M.abscessus_ATCC_199 FAYSRKHSVLGLRVDVEQ---------------------
Mb3281c|M.bovis_AF2122/97 VGYGRRHSAQGLRQARESATRRC----------------
Rv3253c|M.tuberculosis_H37Rv VGYGRRHSAQGLRQARESATRRC----------------
MUL_2597|M.ulcerans_Agy99 VGYGRKHSMVGRRQLAEAAMDK-----------------
MAV_4216|M.avium_104 AGYGYWHSVQGRRDAGEPDRSDALAADPNSPVP------
Mflv_4427|M.gilvum_PYR-GCK LLAKRFGGQGVQDTGDAAPSRSE----------------
MLBr_01305|M.leprae_Br4923 SIRKRVMTIARERMGYTGPFPAIANPPVQPSERSHSQNP