For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GHLDADKKTNHQPIEGDCVSQGEATSVAVEGKGLKGGAVGLLSSVVIGTASTAPAYSLAATLGLIVASRG ELLCGVKAPAIVLLSFIPMYMIAVAYQELNKAEPDCGTTFTWASRAFGPVTGWLGGWGIIAADVIVMANL AQIAGAYSFTFVGELGWQSASNLAHSTLWSTVAGVAWIVVMTCICYRGTDVSARMQYVLLSVELVVLVAL AAVALARVYTNHGDVGSLRPSLSWLWPSGLDFGTQMAPALLITIFIYWGFDTAVACNDETDDPGRTLGRA AVISTFLLLATYAVVTVASIAFAGVGVDGIGLGNPDNSNDVFAAVGPALFGDGVLGKIGLLLLSASILTS ASASTQTTILPTARTTLSMAVYKALPASFAKIHPRYLTPTVSTIAMGAVSILFYVLFTLVSRNLLSALIG SVGLVIAFYYGLTGYACAHYYRATLTKSLRNFVMRGVVPLTGGVVLTVVFIFGLIEYAKRDWLVENGHNV TIFGFGAVAVVGVGSMLLGVVVMLIWWAISPGFFRGRTLRRHSADLLLEPAVSPPAHLALPDSGDMQTVI AADLSNLPAGETAVNPQTGQHLTKDTSGAS*
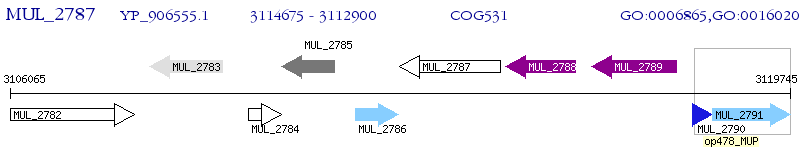
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2787 | - | - | 100% (591) | hypothetical protein MUL_2787 |
| M. ulcerans Agy99 | MUL_1244 | rocE | 4e-13 | 23.12% (372) | cationic amino acid transport integral membrane protein RocE |
| M. ulcerans Agy99 | MUL_2597 | - | 1e-08 | 23.20% (375) | cationic amino acid transport integral membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2022c | - | 5e-27 | 27.21% (441) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_4853 | - | 1e-117 | 45.12% (492) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv1999c | - | 5e-27 | 27.21% (441) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3331c | - | 0.0 | 72.87% (564) | putative amino acid permease |
| M. marinum M | MMAR_3554 | - | 0.0 | 99.15% (591) | hypothetical protein MMAR_3554 |
| M. avium 104 | MAV_2087 | - | 1e-12 | 23.43% (367) | amino acid transporter |
| M. smegmatis MC2 155 | MSMEG_1625 | - | 1e-105 | 40.78% (510) | amino acid transporter, putative |
| M. thermoresistible (build 8) | TH_0471 | - | 1e-10 | 22.25% (409) | POSSIBLE CATIONIC AMINO ACID TRANSPORT INTEGRAL MEMBRANE |
| M. vanbaalenii PYR-1 | Mvan_1574 | - | 1e-123 | 45.63% (504) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2787|M.ulcerans_Agy99 MGHLDADKKTNHQPIEGDCVSQGEATSVAVEGKGLKGGAVGLLSSVVIGT
MMAR_3554|M.marinum_M MGHLDADKKTSHQPIEGDCVSQGEATSVAVEGKGLKGGAVGLLSSVVIGT
MAB_3331c|M.abscessus_ATCC_199 -----------------------MSSDIPVESKGLKGGALGLLSSVVVGL
Mflv_4853|M.gilvum_PYR-GCK ---------------------------------------------MVIGL
Mvan_1574|M.vanbaalenii_PYR-1 ------------------MAITDEPAVPAVGGKGLQAGALGLVGNVVIGL
MSMEG_1625|M.smegmatis_MC2_155 ---MTRSDPQSDPESDPHHALEIVEQSAGLRGKGLAAGKVGTFSGAILGI
Mb2022c|M.bovis_AF2122/97 ----------------------MRRPLDPRDIPDELRRRLGLLDAVVIGL
Rv1999c|M.tuberculosis_H37Rv ----------------------MRRPLDPRDIPDELRRRLGLLDAVVIGL
MAV_2087|M.avium_104 ---------------------MAPGTADH------LRRGIGTFQLTMFGV
TH_0471|M.thermoresistible__bu ------------MTDRLRVKSVEQSIADTDEPDSRLRKDLSWWDLTVFGV
:.*
MUL_2787|M.ulcerans_Agy99 ASTAPAYSLAATLGLIVASRGELLCGVKAPAIVLLSFIPMYMIAVAYQEL
MMAR_3554|M.marinum_M ASTAPAYSLAATLGLIVASRGELLCGVKAPAIVLLSFIPMYMIAVAYQEL
MAB_3331c|M.abscessus_ATCC_199 ASTAPAYSLAATLGLIVASGGALLAGVKAPAIVLISFIPMYLIAVAYQEL
Mflv_4853|M.gilvum_PYR-GCK GAVAPAYSLAATLGYVVLN-----VQEKAPSMFVLAFIPMLLVAFAYKEL
Mvan_1574|M.vanbaalenii_PYR-1 GAVAPAYSLAATLGYVVLN-----VQEKAPAMFVLAFIPMLLVAFAYKEL
MSMEG_1625|M.smegmatis_MC2_155 SSVAPGYTLTASIGLIVAA-----VGLKMPAIFIAGFIPMFLTAYAYREL
Mb2022c|M.bovis_AF2122/97 GSMIGAG-IFAALAPAAY-----AAGSGLLLGLAVAAVVAYCNAISSARL
Rv1999c|M.tuberculosis_H37Rv GSMIGAG-IFAALAPAAY-----AAGSGLLLGLAVAAVVAYCNAISSARL
MAV_2087|M.avium_104 GSTIGTG-IFFVMSQAVP-----EAGPAVIVSFLLAGVAAGLAAVCYAEL
TH_0471|M.thermoresistible__bu SVVIGAG-IFTVTASTAGN----ITGPAISVSFIIAAIACGLAALCYAEF
. : . . . . : * . .:
MUL_2787|M.ulcerans_Agy99 NKAEPDCGTTFTWASRAFGPVTGWLGGWGIIAADVIVMANLAQIAGAYSF
MMAR_3554|M.marinum_M NKAEPDCGTTFTWASRAFGPVTGWLGGWGIIAADVIVMANLAQIAGAYSF
MAB_3331c|M.abscessus_ATCC_199 NKAEPDCGTTFTWASRTFGPIVGWLGGWGIIAADVICLANLAQVAGAYSF
Mflv_4853|M.gilvum_PYR-GCK SQDTPDCGTTFTWATKAFGPWIGWIGGWGLAVSAIIVLANVSEIAAIYLY
Mvan_1574|M.vanbaalenii_PYR-1 SQDTPDCGTTFTWGTKAFGPWIGWIGGWGLAVSAIIVLANVSEIAAIYLY
MSMEG_1625|M.smegmatis_MC2_155 NSRAPDCGASFTWSTKAFGPYVGWMCGWGMVIATIIVLSNLAAIAVEFFY
Mb2022c|M.bovis_AF2122/97 AARYPASGGTYVYGRMRLGDFWGYLAGWGFVVGKTASCAAMALTVG----
Rv1999c|M.tuberculosis_H37Rv AARYPASGGTYVYGRMRLGDFWGYLAGWGFVVGKTASCAAMALTVG----
MAV_2087|M.avium_104 ASAVPVSGSSYSYAYTTLGEVVAMGVAACLLLEYGVATAAVSVNWSGYLN
TH_0471|M.thermoresistible__bu ASTVPVAGSAYTFSYATFGEFVAWIIGWDLILEFSVAAAVVAKGWSSYLG
* .* :: :. :* . . : : ::
MUL_2787|M.ulcerans_Agy99 TFVGELG------WQSASNLAHSTLWSTVAGVAWIVVMTCICYRGTDVSA
MMAR_3554|M.marinum_M TFVGELG------WQSASNLAHSALWSTVAGVAWIVVMTYICYRGTDVSA
MAB_3331c|M.abscessus_ATCC_199 TFVGDLG------WSAASGLADSVLWSTVAGVIWIVIMTYICYRGIEVSA
Mflv_4853|M.gilvum_PYR-GCK NFLG------------LTSLAESVPATVALGCFFIIAMTLVSARGIVVSE
Mvan_1574|M.vanbaalenii_PYR-1 NFLG------------LDSLAESVPATVALGCFFIISMTLVSARGIVVSE
MSMEG_1625|M.smegmatis_MC2_155 LFVARILN-----TPEIADLADNKLVNILTTLVFIALATWVSSRGITTSE
Mb2022c|M.bovis_AF2122/97 -------------------FYVWPAQAHAVAVAVVVALTAVNYAGIQKSA
Rv1999c|M.tuberculosis_H37Rv -------------------FYVWPAQAHAVAVAVVVALTAVNYAGIQKSA
MAV_2087|M.avium_104 KLLSNVVGFQLPHALSAAPWDAQPGYVNLPAVMLIGMCALLLIRGASESA
TH_0471|M.thermoresistible__bu EVFE----------FAGATTRLGPITLDWGALLIIGFVTVLLATGTKLSA
: : : * *
MUL_2787|M.ulcerans_Agy99 RMQYVLLSVELVVLVALAAVALARVYTNHGDVGSLRPSLSWLWP------
MMAR_3554|M.marinum_M RMQYVLLSVELVVLVALAAVALARVYTNHGDVGSLRPSLSWLWP------
MAB_3331c|M.abscessus_ATCC_199 RLQYWLLTVEVVVLIAFAVVALIKVYTHNAESYSLLPSLSWFWP------
Mflv_4853|M.gilvum_PYR-GCK RVQNVLIAIQFGVLITVSIIALVRVFSGTAGAQAVMPEVSWLWP------
Mvan_1574|M.vanbaalenii_PYR-1 RVQNVLIAIQFGVLITVSVIALIRVFTGTAGAQAISPQLSWLWP------
MSMEG_1625|M.smegmatis_MC2_155 HVQYVLVGFQMTVLLAFAITAVIHVGAGNAPEG-LAFDLDWFNPF-----
Mb2022c|M.bovis_AF2122/97 WLTRSIVAVVLVVLTAVVVAAYGSGAADPARLDIG---------------
Rv1999c|M.tuberculosis_H37Rv WLTRSIVAVVLVVLTAVVVAAYGSGAADPARLDIG---------------
MAV_2087|M.avium_104 KVNAIMVMVKLGVLVVFGILAFTAFDMHHLDDFAP---------------
TH_0471|M.thermoresistible__bu NVSLAITVIKVGVVLLVVIVGAFFIKAANYTPFIPPAEAGGEAGRSLDQS
: : . . *: . .
MUL_2787|M.ulcerans_Agy99 ----------SGLDFGTQMAPALLITIFIYWGFDTAVACNDETDDPGRTL
MMAR_3554|M.marinum_M ----------SGLDFGTQMAPALLITIFIYWGFDTAVACNEETDDPGRTP
MAB_3331c|M.abscessus_ATCC_199 ----------GGMDFGTVIAPAVLTTIFIYWGWDTAVACNEESDDPGRTP
Mflv_4853|M.gilvum_PYR-GCK ----------SGLDVSS-IAAAVILCIFIYWGWDACLAVGEETKDPGKTP
Mvan_1574|M.vanbaalenii_PYR-1 ----------SGLDVSS-IAAAVILCIFIYWGWDACLAVGEETKDPGRTP
MSMEG_1625|M.smegmatis_MC2_155 ----------TNLAFGA-FVIGVTGSIFAFWGWDTCLTLGEESKDPTKVP
Mb2022c|M.bovis_AF2122/97 ----------VDAHVWG-MLQAAGLLFFAFAGYARIATLGEEVRDPARTI
Rv1999c|M.tuberculosis_H37Rv ----------VDAHVWG-MLQAAGLLFFAFAGYARIATLGEEVRDPARTI
MAV_2087|M.avium_104 ------------FGVAG-VGTAAGTIFFSYIGLDAVSTAGDEVTNPQKTM
TH_0471|M.thermoresistible__bu LFSLITGAEGSSYGWYG-LLAGASIVFFAFIGFDIIATTAEETKNPQRDV
. . :* : * : :* :* :
MUL_2787|M.ulcerans_Agy99 GRAAVISTFLLLATYAVVTVASIAFAGVGVDGIGLGNPDNSNDVFAAVGP
MMAR_3554|M.marinum_M GRAAVISTFLLLATYAVVTVASIAFAGVGVDGIGLGNPDNSNDVFAAVGP
MAB_3331c|M.abscessus_ATCC_199 GRAAVISTFLLLATYALVSVAAVAFAGVGTEGVGLGNQENAADVFKAIGP
Mflv_4853|M.gilvum_PYR-GCK GIAALITTVILVCTYVLVAYALQSFAGFSEVGIGLNNPNNTDDVLTVLG-
Mvan_1574|M.vanbaalenii_PYR-1 GIAALVTTVILVCTYVLVAFALQSFAGFSEVGIGLNNPNNTDDVLTVLG-
MSMEG_1625|M.smegmatis_MC2_155 GRAGLLCVTTILLTYLLVAVAVMMYAGVGETGLGLGNPENSENVFGALA-
Mb2022c|M.bovis_AF2122/97 PRAIPLALGITLAVYALVAVAVIAVLGP----QRL--ARAAAPLSEAMR-
Rv1999c|M.tuberculosis_H37Rv PRAIPLALGITLAVYALVAVAVIAVLGP----QRL--ARAAAPLSEAMR-
MAV_2087|M.avium_104 PRALIAALLTVTGVYVFVALAALGTQPW----QDFGGQQE-AGLATILD-
TH_0471|M.thermoresistible__bu ARGIMATLGIVTVLYVAVSLVLTGMVHY----TELRAAGENANLATAFA-
. * *: . : : .
MUL_2787|M.ulcerans_Agy99 ALFGDGVLGKIGLLLLSASILTSASASTQTTILPTARTTLSMAVYKALPA
MMAR_3554|M.marinum_M ALFGDGVLGKIGLLLLSASILTSASASTQTTILPTARTTLSMAVYKALPA
MAB_3331c|M.abscessus_ATCC_199 ELFGDSFVGHLGMLLLSASILTSASACTQTTILPTARTTLSMGVYKALPD
Mflv_4853|M.gilvum_PYR-GCK ----EPVAGAIAASALLLTVSVSALSSTQTTILPTARGTLSMAVYEAIPR
Mvan_1574|M.vanbaalenii_PYR-1 ----KPVAGAIAASALLLTVSVSALSSTQTTILPTARGTLSMAVYEAIPK
MSMEG_1625|M.smegmatis_MC2_155 ----DPVLGSWGGPLLFLAVLASSVASLQTTFLPAARAMLAMGAYGAFPK
Mb2022c|M.bovis_AF2122/97 ----VAGVN-WLIPVVQIGAAVAALGSLLALILGVSRTTLAMARDRHLPR
Rv1999c|M.tuberculosis_H37Rv ----VAGVN-WLIPVVQIGAAVAALGSLLALILGVSRTTLAMARDRHLPR
MAV_2087|M.avium_104 ----HVTHGSWASTILAAGAVISIFSVTLVTMYGITRILFAMGRDGLLPP
TH_0471|M.thermoresistible__bu ----ANGID-WAAKVISIGALAGLTTVVIVLVLGQTRVLFAMSRDGLLPR
. : . . . :* ::*. :*
MUL_2787|M.ulcerans_Agy99 SFAKIHPRYLTPTVSTIAMGAVSILFYVLFTLVSRNLLSALIGSVGLVIA
MMAR_3554|M.marinum_M SFAKIHPRYLTPTVSTIAMGAVSILFYVLFTLVSRNLLSALIGSVGLVIA
MAB_3331c|M.abscessus_ATCC_199 SFASIHRTYLTPSTSTIAMGAISIVFYVLFTAISTNLLTALIGSVGLMIA
Mflv_4853|M.gilvum_PYR-GCK RFASVHPRYMTPAFGTIVMGISALFFYLVLKMLSENALLDSVASLGLAVA
Mvan_1574|M.vanbaalenii_PYR-1 RFANVHPRYMTPAFGTIVMGLSALFFYLVLKALSENALLDSVASLGLAVA
MSMEG_1625|M.smegmatis_MC2_155 RFSHVHPRFLVPSFSTLIAGIVTGVFYTVVSLLSEYALLDTIAALGIMIC
Mb2022c|M.bovis_AF2122/97 WLAAVHPRFKVPFRAELVVGAVVAALAATADIRG----AIGFSSFGVLVY
Rv1999c|M.tuberculosis_H37Rv WLAAVHPRFKVPFRAELVVGAVVAALAATADIRG----AIGFSSFGVLVY
MAV_2087|M.avium_104 RFARVNPRTMTPVNNTVIVAVAAATLAAFIPLQN----LADMVSIGTLTA
TH_0471|M.thermoresistible__bu PLSKTGPR-GTPVRITVLVGALVAIAASVFPVGN----LEEMVNIGTLFA
:: .* : . . . .*
MUL_2787|M.ulcerans_Agy99 FYYGLTGYACAHYYRATLTKSLRNFVMRGVVPLTGGVVLTVVFIFGLIEY
MMAR_3554|M.marinum_M FYYGLTGYACAHYYRATLTKSLRNFVMRGVVPLTGGVVLTVVFIFGLIEY
MAB_3331c|M.abscessus_ATCC_199 FYYGLTGFACAWFYRKRLTTSFRDLMMRGVMPLAGGATLTVVFIYGLIQY
Mflv_4853|M.gilvum_PYR-GCK FYYGITAFACVWYFRRTLFSSARNLFFRGVFPLLGGLSMLAAFAISAKDM
Mvan_1574|M.vanbaalenii_PYR-1 FYYGITAFACVWYFRRTLFESTRNLFFRGVFPLLGGLSMLAAFTISAKDM
MSMEG_1625|M.smegmatis_MC2_155 WYYGITAFACIWYFRRELFTDAHNIVFKFLFPLIGGLVLAAVFGFSLKES
Mb2022c|M.bovis_AF2122/97 YAIANASALTLGLDEGRPRR---------LIPLVG-LIGCVVLAFALPLS
Rv1999c|M.tuberculosis_H37Rv YAIANASALTLGLDEGRPRR---------LIPLVG-LIGCVVLAFALPLS
MAV_2087|M.avium_104 FVVVSVGVIVLRVREPDLPRGFR----VPGYPVTP-VLSIMACGYILASL
TH_0471|M.thermoresistible__bu FALVSAGVIVLRRTRPDLKRGFR----VPFVPWLP-LAAIAACLWLMLNL
: .. . * .
MUL_2787|M.ulcerans_Agy99 AKRDWLVEN-GHNVTIFG-FGAVAVVGVGSMLLGVVVMLIWWAISPG-FF
MMAR_3554|M.marinum_M AKRDWLVEN-GHNVTIFG-FGAVAVVGVGSMLLGVVVMLIWWAISPG-FF
MAB_3331c|M.abscessus_ATCC_199 AKPNWLIDDDGNNVTVFG-VGAVAVVGVGALVLGVVLMMLWRVAAPP-FF
Mflv_4853|M.gilvum_PYR-GCK IQPDY-------GHTAFGSIGGVFVLGVGMLVLGIPLMLACFAFGTKRFF
Mvan_1574|M.vanbaalenii_PYR-1 IQPDY-------GYTAFGPVGGVFVLGVGMLVLGIPLMLACFAFGTKRFF
MSMEG_1625|M.smegmatis_MC2_155 MDPEN------GSGAELGGIGLVFYIGFGILLLGAVLMLVMRTRSPE-FF
Mb2022c|M.bovis_AF2122/97 SVAAG-----------------AAVLGVGVAAYGVRRIITRRARQTDSGD
Rv1999c|M.tuberculosis_H37Rv SVAAG-----------------AAVLGVGVAAYGVRRIITRRARQTDSGD
MAV_2087|M.avium_104 HWYTW-----------------IAFSGWVLLALIFYFVWGRHHSALNDAA
TH_0471|M.thermoresistible__bu TGLTW-----------------VRFLGWMALGLVLYFFYGRHHSMLGRGM
* .
MUL_2787|M.ulcerans_Agy99 RGRTLRRHSADLLLEPAVSPPAHLALPDSGD-MQTVIAADLSNLPAGETA
MMAR_3554|M.marinum_M RGRTLRRHSADLLLEPAVSPPAHLALPDSGD-MQTVIAADLSNLPAGETA
MAB_3331c|M.abscessus_ATCC_199 QGQTLPRRSADLVLAPAAAT-THFGLPDAVEAQQTVIAGDLSNLPEGEVA
Mflv_4853|M.gilvum_PYR-GCK RGETLN-------------ADTAVKVPDTF--------------------
Mvan_1574|M.vanbaalenii_PYR-1 RGETLN-------------ADTEVKVPDTF--------------------
MSMEG_1625|M.smegmatis_MC2_155 RGETLSR--------------NTPVIPG----------------------
Mb2022c|M.bovis_AF2122/97 TQRSGHPSAT----------------------------------------
Rv1999c|M.tuberculosis_H37Rv TQRSGHPSAT----------------------------------------
MAV_2087|M.avium_104 VDPSGQER------------------------------------------
TH_0471|M.thermoresistible__bu TDISARRTG-----------------------------------------
:
MUL_2787|M.ulcerans_Agy99 VNPQTGQHLTKDTSGAS
MMAR_3554|M.marinum_M VNPQTGQHLTKDTSGAS
MAB_3331c|M.abscessus_ATCC_199 VNPQTGEEFTKD-----
Mflv_4853|M.gilvum_PYR-GCK -----------------
Mvan_1574|M.vanbaalenii_PYR-1 -----------------
MSMEG_1625|M.smegmatis_MC2_155 -----------------
Mb2022c|M.bovis_AF2122/97 -----------------
Rv1999c|M.tuberculosis_H37Rv -----------------
MAV_2087|M.avium_104 -----------------
TH_0471|M.thermoresistible__bu -----------------