For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAPGTADHLRRGIGTFQLTMFGVGSTIGTGIFFVMSQAVPEAGPAVIVSFLLAGVAAGLAAVCYAELASA VPVSGSSYSYAYTTLGEVVAMGVAACLLLEYGVATAAVSVNWSGYLNKLLSNVVGFQLPHALSAAPWDAQ PGYVNLPAVMLIGMCALLLIRGASESAKVNAIMVMVKLGVLVVFGILAFTAFDMHHLDDFAPFGVAGVGT AAGTIFFSYIGLDAVSTAGDEVTNPQKTMPRALIAALLTVTGVYVFVALAALGTQPWQDFGGQQEAGLAT ILDHVTHGSWASTILAAGAVISIFSVTLVTMYGITRILFAMGRDGLLPPRFARVNPRTMTPVNNTVIVAV AAATLAAFIPLQNLADMVSIGTLTAFVVVSVGVIVLRVREPDLPRGFRVPGYPVTPVLSIMACGYILASL HWYTWIAFSGWVLLALIFYFVWGRHHSALNDAAVDPSGQER
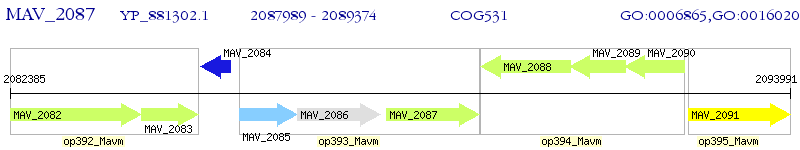
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2087 | - | - | 100% (461) | amino acid transporter |
| M. avium 104 | MAV_4216 | - | 1e-72 | 35.53% (470) | cationic amino acid transporter |
| M. avium 104 | MAV_4626 | - | 7e-22 | 23.29% (468) | gaba permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2347c | rocE | 0.0 | 81.60% (451) | cationic amino acid transport integral membrane protein RocE |
| M. gilvum PYR-GCK | Mflv_4722 | - | 1e-82 | 37.63% (473) | amino acid permease-associated region |
| M. tuberculosis H37Rv | Rv2320c | rocE | 0.0 | 81.60% (451) | cationic amino acid transport integral membrane protein RocE |
| M. leprae Br4923 | MLBr_01974 | - | 4e-25 | 23.51% (404) | putative permease |
| M. abscessus ATCC 19977 | MAB_3842 | - | 0.0 | 71.15% (461) | cationic amino acid transport integral membrane protein |
| M. marinum M | MMAR_3621 | rocE | 0.0 | 78.16% (467) | cationic amino acid transport integral membrane protein RocE |
| M. smegmatis MC2 155 | MSMEG_1412 | - | 1e-176 | 67.84% (454) | amino acid permease |
| M. thermoresistible (build 8) | TH_0471 | - | 4e-83 | 37.42% (481) | POSSIBLE CATIONIC AMINO ACID TRANSPORT INTEGRAL MEMBRANE |
| M. ulcerans Agy99 | MUL_1244 | rocE | 0.0 | 77.73% (467) | cationic amino acid transport integral membrane protein RocE |
| M. vanbaalenii PYR-1 | Mvan_1740 | - | 7e-82 | 37.00% (473) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4722|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1740|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0471|M.thermoresistible__bu --------------------------------------------------
MMAR_3621|M.marinum_M --------------------------------------------------
MUL_1244|M.ulcerans_Agy99 --------------------------------------------------
Mb2347c|M.bovis_AF2122/97 --------------------------------------------------
Rv2320c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2087|M.avium_104 --------------------------------------------------
MAB_3842|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1412|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_01974|M.leprae_Br4923 MFLSADVSCIEIVDINCSELRNDRRIPPQLALFCTNAANKAESAVTSHGQ
Mflv_4722|M.gilvum_PYR-GCK --------------------------MTQEPVTQAPGRLRLKSVEQSIAD
Mvan_1740|M.vanbaalenii_PYR-1 --------------------------MVQEPGAQTVGRRRTKSVEQSIAD
TH_0471|M.thermoresistible__bu ----------------------------------MTDRLRVKSVEQSIAD
MMAR_3621|M.marinum_M --------------------------MPATSIRLREQMLRRRPVGGAPVA
MUL_1244|M.ulcerans_Agy99 --------------------------MPATSIRLREQMLRRRPAGGAPVA
Mb2347c|M.bovis_AF2122/97 --------------------------MPTTSMSLRELMLRRRPVSGAPVA
Rv2320c|M.tuberculosis_H37Rv --------------------------MPTTSMSLRELMLRRRPVSGAPVA
MAV_2087|M.avium_104 ------------------------------------------------MA
MAB_3842|M.abscessus_ATCC_1997 --------------------------MAPSSPSLAQQLLRRRPTSGAPTA
MSMEG_1412|M.smegmatis_MC2_155 --------------------------MSAPPTSMAGQMMRRRPVIGAPVA
MLBr_01974|M.leprae_Br4923 LPIMANRRWAPHTRRQSGRATSTCSSLMRFSASESRAHSRFECYTLVRWS
Mflv_4722|M.gilvum_PYR-GCK TDEPGTKLRKDLNWWDLTVFGVSVVIGAGIFTITASTAANITGPAISLSF
Mvan_1740|M.vanbaalenii_PYR-1 TDEPSTRLRKELNWWDLTVFGVSVVIGAGIFTITASTAANITGPAISLSF
TH_0471|M.thermoresistible__bu TDEPDSRLRKDLSWWDLTVFGVSVVIGAGIFTVTASTAGNITGPAISVSF
MMAR_3621|M.marinum_M EGAAG-SLKRSFGTFELTMFGVGSTVGTGIFFVLAEAVP-EAGPAVIVSF
MUL_1244|M.ulcerans_Agy99 EGAAG-SLKRSFGTFELTMFGVGSTVGTGIFFVLAEAVP-EAGPAVIVSF
Mb2347c|M.bovis_AF2122/97 SGASG-NLKRSFGTFQLTMFGVGATIGTGIFFVLAQAVP-EAGPGVIVSF
Rv2320c|M.tuberculosis_H37Rv SGASG-NLKRSFGTFQLTMFGVGATIGTGIFFVLAQAVP-EAGPGVIVSF
MAV_2087|M.avium_104 PGTAD-HLRRGIGTFQLTMFGVGSTIGTGIFFVMSQAVP-EAGPAVIVSF
MAB_3842|M.abscessus_ATCC_1997 HGAGG-HLKQSMGTFQLTMIGVGATVGTGIFFILAESVP-KAGPAVMASF
MSMEG_1412|M.smegmatis_MC2_155 HGASD-HLKRSIGTFQLTLFGVGATVGTGIFLVLQEAVP-EAGPAVLVSF
MLBr_01974|M.leprae_Br4923 IRGQGGRAISKLGFWSVVMLGINSIIGAGIFMTPGEVIK-IAGPFAPIAY
. :. :.:.::*:. :*:*** . :** ::
Mflv_4722|M.gilvum_PYR-GCK IFAAIACGLAALCYAEFASTVPVAGSAYTFSYATFGEFVAWIIGWDLILE
Mvan_1740|M.vanbaalenii_PYR-1 IIAAIACGLAALCYAEFASTVPVAGSAYTFSYATFGEFVAWIIGWDLILE
TH_0471|M.thermoresistible__bu IIAAIACGLAALCYAEFASTVPVAGSAYTFSYATFGEFVAWIIGWDLILE
MMAR_3621|M.marinum_M IIAGVAAGLAAICYAELASAVPVSGSSYSYAYATLGEAVAMGVASCLLLE
MUL_1244|M.ulcerans_Agy99 IIAGVAAGLAAICCAELASAVPVSGSSYSYAYATLGEAVAMGVASCLLLE
Mb2347c|M.bovis_AF2122/97 IIAGIAAGLAAICYAELASAVPISGSAYSYAYTTLGEAVAMVVAACLLLE
Rv2320c|M.tuberculosis_H37Rv IIAGIAAGLAAICYAELASAVPISGSAYSYAYTTLGEAVAMVVAACLLLE
MAV_2087|M.avium_104 LLAGVAAGLAAVCYAELASAVPVSGSSYSYAYTTLGEVVAMGVAACLLLE
MAB_3842|M.abscessus_ATCC_1997 LVAGLAAGLAALCYAELASAVPVSGSSFSYAYTTLGELAAMVVAACLLLE
MSMEG_1412|M.smegmatis_MC2_155 VLAGVAAGLSALCYAEMASAVPVSGSTYSYAYTTMGEFVAMGVAACLLLE
MLBr_01974|M.leprae_Br4923 TLAGIFAAVLAIVFATAAKYVKTNGSSYAYTTAAFGHRIGIYVGVTHAIT
.*.: ..: *: * *. * **::::: :::*. . :. :
Mflv_4722|M.gilvum_PYR-GCK FAVAAAVVAKGWSSYLG----TVFGFAG-GTAEVAGFD----------LD
Mvan_1740|M.vanbaalenii_PYR-1 FAVAAAVVAKGWSSYLG----TVFGFAG-GTTEVAGFQ----------LD
TH_0471|M.thermoresistible__bu FSVAAAVVAKGWSSYLG----EVFEFAG-ATTRLGPIT----------LD
MMAR_3621|M.marinum_M YGVATAAVAVGWSGYVNKLLHNVFGFQLPQALSEAPWD-ERPGEAAGYLN
MUL_1244|M.ulcerans_Agy99 YGVATAAVAVGLSGYVNKLLHNVFGFQLPQALSEAPWD-ERPGEAAGYLN
Mb2347c|M.bovis_AF2122/97 YGVATAAVAVGWSGYVNKLLSNLFGFQMPHVLSAAPWD-THPG----WVN
Rv2320c|M.tuberculosis_H37Rv YGVATAAVAVGWSGYVNKLLSNLFGFQMPHVLSAAPWD-THPG----WVN
MAV_2087|M.avium_104 YGVATAAVSVNWSGYLNKLLSNVVGFQLPHALSAAPWD-AQPG----YVN
MAB_3842|M.abscessus_ATCC_1997 YGVSAAAVAVGWSQYLDKLLVNLFGWHLPHAISAAPWDSADPAAPHPWLN
MSMEG_1412|M.smegmatis_MC2_155 YGVSMSATSIGWGGYLNQLLDDVFGFRIPQALTSAPWG-ENPG----IMN
MLBr_01974|M.leprae_Br4923 ASIAWGVLASSFVSTLLRVAFPEKIWADNDELISV-------------KT
.:: .. : . : :
Mflv_4722|M.gilvum_PYR-GCK WGALVIIAIVTAILAMGTKLSASVSLAITALKVSVVLLVVIVGAFYIKME
Mvan_1740|M.vanbaalenii_PYR-1 WGALVIIALVTLILVMGTKLSAGVSLAITAVKVSVVLLVVIVGAFYIKAE
TH_0471|M.thermoresistible__bu WGALLIIGFVTVLLATGTKLSANVSLAITVIKVGVVLLVVIVGAFFIKAA
MMAR_3621|M.marinum_M LPAIILIGLCALLLIRGVSESAKVNAIMVLIKLGVLGMFVILALTAYDTN
MUL_1244|M.ulcerans_Agy99 LPAIILIGLCALLLIRGVSESAKVNAIMVLIKLGVLGMFVILALTAYDTN
Mb2347c|M.bovis_AF2122/97 LPAVILIGLCALLLIRGASESARVNAIMVLIKLGVLGMFMIIAFSAYSAD
Rv2320c|M.tuberculosis_H37Rv LPAVILIGLCALLLIRGASESARVNAIMVLIKLGVLGMFMIIAFSAYSAD
MAV_2087|M.avium_104 LPAVMLIGMCALLLIRGASESAKVNAIMVMVKLGVLVVFGILAFTAFDMH
MAB_3842|M.abscessus_ATCC_1997 LPAVLLIIMCAILLIRGTSESALVNTVMVLLKLAVLSVFAIIAFTAFHAG
MSMEG_1412|M.smegmatis_MC2_155 LTATLLIVMCGLLLIRGASESALVNTIMVIIKLAVLALFVAVAFTAFTTD
MLBr_01974|M.leprae_Br4923 LTFLVFIAVLLAINIFGNLVIRWANGISALGKIFALSVFIAGGLWTIITQ
::* . : * .. . *: .: :. .
Mflv_4722|M.gilvum_PYR-GCK NFSPFIPAPEPGEAGGTGAEQSLFSLMTGAEGSHYGWYGLLAGASIVFFA
Mvan_1740|M.vanbaalenii_PYR-1 NFSPFVPAPEEGGSGGTGTEQSLFSLLTGAEGSHYGWYGVLAGASIVFFA
TH_0471|M.thermoresistible__bu NYTPFIPPAEAGGEAGRSLDQSLFSLITGAEGSSYGWYGLLAGASIVFFA
MMAR_3621|M.marinum_M HLKDFAP---------------------------FGVAGVGAAAGTIFFS
MUL_1244|M.ulcerans_Agy99 HLKDFAP---------------------------FGVAGVGAAAGTIFFS
Mb2347c|M.bovis_AF2122/97 HLKDFVP---------------------------FGVAGIGSAAGTIFFS
Rv2320c|M.tuberculosis_H37Rv HLKDFVP---------------------------FGVAGIGSAAGTIFFS
MAV_2087|M.avium_104 HLDDFAP---------------------------FGVAGVGTAAGTIFFS
MAB_3842|M.abscessus_ATCC_1997 NFSDFAP---------------------------FGVSGIGAAAGTIFFT
MSMEG_1412|M.smegmatis_MC2_155 HFAGFWD---------------------------KGFAGITAAAGSIFFT
MLBr_01974|M.leprae_Br4923 HVNNYVTSR-------SAYQPTTYSLLGVTEIGKSTAASMTLATIVALYA
: : .: .: :::
Mflv_4722|M.gilvum_PYR-GCK FIGFDIVATTAEETKVPQRDVPRGILASLGIVTVLYVAVAIVLSGMVSYK
Mvan_1740|M.vanbaalenii_PYR-1 FIGFDIVATTAEETRNPQRDVPRGILASLAIVTVLYVAVAVVLTGMVSYT
TH_0471|M.thermoresistible__bu FIGFDIIATTAEETKNPQRDVARGIMATLGIVTVLYVAVSLVLTGMVHYT
MMAR_3621|M.marinum_M FIGLDAVSTAGEEVRNPQQSVPRALIAALFVVTGVYVLVAFAALGTQPWQ
MUL_1244|M.ulcerans_Agy99 FIGLDAVSTAGEEVRNPQQSVPRALIAALFVVTGVYVLVAFAALGTQPWQ
Mb2347c|M.bovis_AF2122/97 YIGLDAVSTAGDEVKDPQKTMPRALIAALVVVTGVYVLVALAALGTQPWQ
Rv2320c|M.tuberculosis_H37Rv YIGLDAVSTAGDEVKDPQKTMPRALIAALVVVTGVYVLVALAALGTQPWQ
MAV_2087|M.avium_104 YIGLDAVSTAGDEVTNPQKTMPRALIAALLTVTGVYVFVALAALGTQPWQ
MAB_3842|M.abscessus_ATCC_1997 YVGLDTVSTAGEEVKDPQRTMPRALIAALLIVTAFYLVVALAALGAQPWK
MSMEG_1412|M.smegmatis_MC2_155 FIGLDAVSTAGDEVKNPQKTMPRAILGALVVVASVYILVAFAGLGTQSAD
MLBr_01974|M.leprae_Br4923 FTGFESIANAAEEMNTPDRTLPKAIPLTILTVGVIYILALVVAMLLGSDK
: *:: ::.:.:* *:: :.:.: :: * .*: . ..
Mflv_4722|M.gilvum_PYR-GCK DL-RDA-ESPNLATAFS--LNGVDWAAKVISIGALAGLTTVVIVLVLGQT
Mvan_1740|M.vanbaalenii_PYR-1 EL-RDA-ETQNLATAFS--LNGVDWAAKVISIGALAGLTTVVIVLVLGQT
TH_0471|M.thermoresistible__bu EL-RAAGENANLATAFA--ANGIDWAAKVISIGALAGLTTVVIVLVLGQT
MMAR_3621|M.marinum_M MF--AGQEEAGLATILDNVTHS-AWASTVLAMGAVISIFTVTLVTMYGQT
MUL_1244|M.ulcerans_Agy99 MF--AGQEEAGLATILDNVTHS-AWASTVLAMGAVISIFTVTLVTMYGQT
Mb2347c|M.bovis_AF2122/97 DF--AEQETAGLAIILDNVTHG-EWASTILAAGAVVSIFTVTLVTMYGQT
Rv2320c|M.tuberculosis_H37Rv DF--AEQETAGLAIILDNVTHG-EWASTILAAGAVVSIFTVTLVTMYGQT
MAV_2087|M.avium_104 DF--GGQQEAGLATILDHVTHG-SWASTILAAGAVISIFSVTLVTMYGIT
MAB_3842|M.abscessus_ATCC_1997 EF--EGQ-SAGLALILDKVTGS-TSASTFLAAGAVISIFSITLVSMYGQT
MSMEG_1412|M.smegmatis_MC2_155 EFGSEEQSEAGLAVILTNILHGQTWASTILSFGAVISIFSVTLVVMYGQT
MLBr_01974|M.leprae_Br4923 IV--ATNHTVKLAAAVGNNTFR-----TIIVVGALVSMFGINVAASFGAP
. ** . ..: **: .: : :. * .
Mflv_4722|M.gilvum_PYR-GCK RVLFAMSRDGLLPRQLAKTGEH-GTPVRITLLVGAVVAVTATVFPID--K
Mvan_1740|M.vanbaalenii_PYR-1 RVLFAMSRDGLLPRQLAKTGEH-GTPVRITLIVGAVVAVTATVFPIG--K
TH_0471|M.thermoresistible__bu RVLFAMSRDGLLPRPLSKTGPR-GTPVRITVLVGALVAIAASVFPVG--N
MMAR_3621|M.marinum_M RILFAMGRDGLLPARFAAVHPRFQTPVSNTVVVAIAAAVLAALVPLN--R
MUL_1244|M.ulcerans_Agy99 RILFAMGRDGLLPARFAAVHPRFQTPVSNTVVVAIAAAVLAALVPLN--R
Mb2347c|M.bovis_AF2122/97 RILFAMGRDGLLPARFAKVNPRTMTPVHNTVIVAIFASTLAAFIPLD--S
Rv2320c|M.tuberculosis_H37Rv RILFAMGRDGLLPARFAKVNPRTMTPVHNTVIVAIFASTLAAFIPLD--S
MAV_2087|M.avium_104 RILFAMGRDGLLPPRFARVNPRTMTPVNNTVIVAVAAATLAAFIPLQ--N
MAB_3842|M.abscessus_ATCC_1997 RILFAIGRDGLLPSVFAQVNPRTLTPVNNTIIVAVVVGFMAAFVPLD--S
MSMEG_1412|M.smegmatis_MC2_155 RILFAMGRDGLLPSMFARVNSHTMTPINNTIVVALVTGTLAGFVPLD--Y
MLBr_01974|M.leprae_Br4923 RLWTALSDGRILPTRLS-SKNRFGVPIVAFGITAFLTLAFPLSLRFDSLN
*: *:. . :** :: : .*: :.. . . . .
Mflv_4722|M.gilvum_PYR-GCK LEEMVNIGTLFAFVLVSAGVIVLRRTRPDLKRGFRTPWVP--VLPILSII
Mvan_1740|M.vanbaalenii_PYR-1 LEEMVNIGTLFAFVLVSAGVLVLRRTRPDLKRGFRTPWVP--LLPILAIA
TH_0471|M.thermoresistible__bu LEEMVNIGTLFAFALVSAGVIVLRRTRPDLKRGFRVPFVP--WLPLAAIA
MMAR_3621|M.marinum_M LADMVSIGTLTAFIVVSVGVIILRVREPALPRGFWVPGYP--VTPILSIL
MUL_1244|M.ulcerans_Agy99 LADMVSIGTLTAFIVVSVGVIILRVREPALPRGFWVPGYP--VTPILSIL
Mb2347c|M.bovis_AF2122/97 LADMVSIGTLTAFSVVAVGVIVLRVREPDLPRGFKVPGYP--VTPVLSVL
Rv2320c|M.tuberculosis_H37Rv LADMVSIGTLTAFSVVAVGVIVLRVREPDLPRGFKVPGYP--VTPVLSVL
MAV_2087|M.avium_104 LADMVSIGTLTAFVVVSVGVIVLRVREPDLPRGFRVPGYP--VTPVLSIM
MAB_3842|M.abscessus_ATCC_1997 LANMVSIGTLTAFIVVSIGVIILRVREPELPRAFRVPGYP--VTPVLSVI
MSMEG_1412|M.smegmatis_MC2_155 LWDLVSIGTLVAFIVVSAGVVLLRVREPDLPRSFKVPGYP--VTPVLSII
MLBr_01974|M.leprae_Br4923 LTGLAVIARFIQFIIVPIALFALARSRTTNSVYVKRNVFTDKVLPITAVV
* :. *. : * :*. .:. * .. . . *: ::
Mflv_4722|M.gilvum_PYR-GCK ACVWLMLNLTGLTWIRFSV---WMAIGVVLYFVYGRRHSLVA---RRQRE
Mvan_1740|M.vanbaalenii_PYR-1 ACVWLMLNLTGLTWIRFLI---WMAIGVVVYFLYGRRHSLVG---RRMG-
TH_0471|M.thermoresistible__bu ACLWLMLNLTGLTWVRFLG---WMALGLVLYFFYGRHHSMLG---RGMTD
MMAR_3621|M.marinum_M ACGYLLYSLHWYTWIAFGG---WVGIALIFYLFWGRHHSRLN---APALH
MUL_1244|M.ulcerans_Agy99 ACGYLLYSLHWYTWIAFGG---WVGIALIFYLFWGRHHSRLN---APALH
Mb2347c|M.bovis_AF2122/97 ACGYILASLHWYTWLAFSG---WVAVAVIFYLMWGRHHSALN---EEVP-
Rv2320c|M.tuberculosis_H37Rv ACGYILASLHWYTWLAFSG---WVAVAVIFYLMWGRHHSALN---EEVP-
MAV_2087|M.avium_104 ACGYILASLHWYTWIAFSG---WVLLALIFYFVWGRHHSALN---DAAVD
MAB_3842|M.abscessus_ATCC_1997 ACGYILFSLPWITWIAFGA---WVAIVLVFYLVWGRHHSALN---DLPSD
MSMEG_1412|M.smegmatis_MC2_155 ACLFVLYGLPAVTWLWFSV---WVGAVLLFYLLWGRRHSALNDGGDGYIP
MLBr_01974|M.leprae_Br4923 VSAGLMVSFDYRSIFLTRGGPNYFSISLIAITFIGIPAVAYLHYYRTSGI
.. :: .: : . :. :: . *
Mflv_4722|M.gilvum_PYR-GCK RAGLHTDTP---------
Mvan_1740|M.vanbaalenii_PYR-1 ------------------
TH_0471|M.thermoresistible__bu ISARRTG-----------
MMAR_3621|M.marinum_M DNPTGEER----------
MUL_1244|M.ulcerans_Agy99 DNPTGEER----------
Mb2347c|M.bovis_AF2122/97 ------------------
Rv2320c|M.tuberculosis_H37Rv ------------------
MAV_2087|M.avium_104 --PSGQER----------
MAB_3842|M.abscessus_ATCC_1997 PAELSEKEPV--------
MSMEG_1412|M.smegmatis_MC2_155 GAAAGEDDVMTGHPEDRA
MLBr_01974|M.leprae_Br4923 K-----------------