For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDEALVAVVTGASRGAGRGIAAALAARGWRVYATSRSVADAPPGGVAVRVDHRDDTATAALFERVQQESG RLDLLVNNAAAISDDLVDPKPFWEKPLALADVLDVGLRSSYVASWYAAPLLVAGGRGLIAFTSSPGSVCY MHGPAYGAQKAGIDKMAADMAVDFHDTGVATVSIWMGILLTEKLRGAFGQHPDGQLALAKTAEHAETPEF TGYLIDALYRDPALGELSGQTVIGAELAERYGITDEGGRRPPSHRHMLGAPRTPSTVVVR*
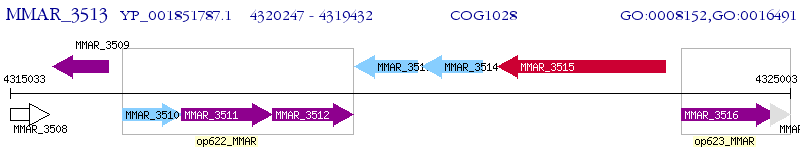
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3513 | - | - | 100% (271) | oxidoreductase |
| M. marinum M | MMAR_0295 | - | 2e-12 | 30.73% (192) | short-chain type dehydrogenase/reductase |
| M. marinum M | MMAR_4756 | - | 2e-11 | 32.26% (186) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3579c | - | 4e-12 | 28.72% (195) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2659 | - | 1e-113 | 76.60% (265) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3549c | - | 4e-12 | 28.72% (195) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 4e-11 | 26.21% (206) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0952 | - | 4e-12 | 30.42% (240) | short chain dehydrogenase |
| M. avium 104 | MAV_3035 | - | 1e-126 | 84.53% (265) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_2923 | - | 1e-118 | 80.38% (265) | dehydrogenase/reductase SDR family protein member 1 |
| M. thermoresistible (build 8) | TH_1294 | - | 1e-108 | 72.69% (271) | dehydrogenase/reductase SDR family protein member 1 |
| M. ulcerans Agy99 | MUL_2755 | - | 1e-115 | 98.06% (206) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_3919 | - | 1e-116 | 76.38% (271) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3513|M.marinum_M ------------MSDEA----LVAVVTGASRGAGRGIAAALAARGWRVYA
MUL_2755|M.ulcerans_Agy99 --------------------------------------------------
MAV_3035|M.avium_104 ----------------------MAVVTGASRGAGRGISAALADRGWRVYA
MSMEG_2923|M.smegmatis_MC2_155 --------------MDT----PVAVVTGASRGAGRGIAAALLAAGWKVYV
TH_1294|M.thermoresistible__bu ------------VTSEA----PVAVVTGASRGAGRGIARALLSRGWRVYG
Mflv_2659|M.gilvum_PYR-GCK ------------MSDIAPVTGPVAVVTGAGRGAGLGIATALAAHGWRVYG
Mvan_3919|M.vanbaalenii_PYR-1 ------------MGDIA----PVAVVTGASRGAGQGIATALAGRGWRVYG
Mb3579c|M.bovis_AF2122/97 -----MTLAEAADAINFGLAGRVVLVTGGVRGVGAGISSVFAEQGATVIT
Rv3549c|M.tuberculosis_H37Rv -----MTLAEAADAINFGLAGRVVLVTGGVRGVGAGISSVFAEQGATVIT
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
MAB_0952|M.abscessus_ATCC_1997 --MVVESAEPAERKVMFDLSARSVLVTGGTKGIGRGIATVFARAGANVAV
MMAR_3513|M.marinum_M TSRSVA-----------DAPPGGVAVRVDHRDDTATAALFERVQQESGRL
MUL_2755|M.ulcerans_Agy99 ------------------------------------------MQQESGRL
MAV_3035|M.avium_104 TSRTIA-----------DTPSGVVPVRVDHRDDAAVSDLFDRVRRESGRL
MSMEG_2923|M.smegmatis_MC2_155 TGRTVT-----------DPGPGGVAVPVDHSDDVAVGALFRRVADESGRL
TH_1294|M.thermoresistible__bu TGRTVT-----------DAD-GVIPAPLDHADDSAVAALFDRVAAECGRL
Mflv_2659|M.gilvum_PYR-GCK TGRGIA-----------EDPAWGTGIRVDHRDDDAVGRVFDRVGAEAGRL
Mvan_3919|M.vanbaalenii_PYR-1 TGRTVA-----------DAPAWGTGVRVDHRDDDAVGALFERVGAEAGRL
Mb3579c|M.bovis_AF2122/97 CARRA------------VDGQPYEFHRCDIRDEDSVKRLVGEIGERHGRL
Rv3549c|M.tuberculosis_H37Rv CARRA------------VDGQPYEFHRCDIRDEDSVKRLVGEIGERHGRL
MLBr_01807|M.leprae_Br4923 THRAS------------VVPDWFFGVECDVTDNDAIGAAFKAVEEHQGPV
MAB_0952|M.abscessus_ATCC_1997 AARSPRELSSVTAELGELGAGNVIGVRLDVSDPGSCADAARTVVDAFGAL
: * :
MMAR_3513|M.marinum_M DLLVNNAAAISDDLVDPKPFWEKPLALADVLDVGLRSSYVASWYAAPLLV
MUL_2755|M.ulcerans_Agy99 DLLVNNAAAISDDLVDPKPFWEKPLALADVLDVGLRSSYVASWYAAPLLV
MAV_3035|M.avium_104 DLLVNNAATISDNLVSSKPFWEKPLDLADVLDVGLRSSYVASWYAAPLLV
MSMEG_2923|M.smegmatis_MC2_155 DLLVNNAAAIHDALVDPKPFWEKPIELADVLDVGLRSAYVASWHAAPLLL
TH_1294|M.thermoresistible__bu DLLVNNAAAVHPALTDPRPFWQKSVELADILDIGLRSAYVAAYHAAPLLL
Mflv_2659|M.gilvum_PYR-GCK DLLVNNAAAISDNLVSAAPFWEKPRDLADVLTVGLRSSYIASWYAAPLLA
Mvan_3919|M.vanbaalenii_PYR-1 DLLVNNAAVISDDLVSPKPFWEKPRALADVLDVGLRSAYIASWHAAPLLT
Mb3579c|M.bovis_AF2122/97 DMLVNNAGGSPYALAAE----ATHNFHRKIVELNVLAPLLVSQHANVLMQ
Rv3549c|M.tuberculosis_H37Rv DMLVNNAGGSPYALAAE----ATHNFHRKIVELNVLAPLLVSQHANVLMQ
MLBr_01807|M.leprae_Br4923 EVLVANAGITSDMLIMR----MSEEQFQKVIDVNLTGVFRVAKLASRNMI
MAB_0952|M.abscessus_ATCC_1997 DVVCANAGIFPEARLDT----MTPEQLSEVLDVNVKGTVYTVQACLAPLT
::: **. . .:: :.: . . . :
MMAR_3513|M.marinum_M AGGRG-LIAFTSSPGSVCYMHG-PAYGAQKAGIDKMAADMAVDFHDTGVA
MUL_2755|M.ulcerans_Agy99 AGGRG-LIAFTSSPGSVCYMHG-PAYGAQKAGIDKMAADMAVDFHDTGVA
MAV_3035|M.avium_104 AGGRG-LIAFTSSPGSVCYMHG-PAYGAQKAGVDKMAADMAVDFRGTGVA
MSMEG_2923|M.smegmatis_MC2_155 AGDKG-LIAFTSSPGSVCYMHG-PAYGAQKAGVDKLAADMAVDFRGTSVC
TH_1294|M.thermoresistible__bu AADRA-LIAFTSSPGSVCYMHG-PAYGAQKAGIDKMAADMAVDFRGTPVC
Mflv_2659|M.gilvum_PYR-GCK AQDRA-LIAFTSSPGSVCYMHG-PAYGAQKAGVDKMAADMAVDFRGTGVA
Mvan_3919|M.vanbaalenii_PYR-1 AQQRG-LVVFTSSPGSVCYMHG-PAYGAQKAGVDKMAADMAVDFRGTGVS
Mb3579c|M.bovis_AF2122/97 AQPNGGSIVNICSVSGRRPTPGTAAYGAAKAGLENLTTTLAVEWAPK-VR
Rv3549c|M.tuberculosis_H37Rv AQPNGGSIVNICSVSGRRPTPGTAAYGAAKAGLENLTTTLAVEWAPK-VR
MLBr_01807|M.leprae_Br4923 KQRFGRYIFMGSVVG-LSGVPGQANYAASKSGLIGMARSLARELGSRSIT
MAB_0952|M.abscessus_ATCC_1997 ASGRGRVILTSSITGPVTGYPGWSHYGASKAAQLGFMRTAAIELAPRGVT
. : . . * . *.* *:. : * : :
MMAR_3513|M.marinum_M TVSIWMGILLTEKLRGAFGQHPDGQLALAKTAEHAETPEFTGYLIDALYR
MUL_2755|M.ulcerans_Agy99 TVSIWMGILLTEKLRGAFGQHPDGQRTLAKTAEHAETPEFTGYLIGALYR
MAV_3035|M.avium_104 TVSIWMGILLTEKLRAAFGADP---AALAATAEHAETPEFTGYVIDALFD
MSMEG_2923|M.smegmatis_MC2_155 TVSIWMGILLTEKLRAAFAGNP---EALAETAKHAETPEFTGRLIDALYR
TH_1294|M.thermoresistible__bu TVSIWMGILLTEQMRRAFTGRP---DALAAMAEHAETPEFTGHILDALYR
Mflv_2659|M.gilvum_PYR-GCK TVSVWMGILLTAKLRGAFDSDP---DALAAFAAHAETPEFTGHVIDALFR
Mvan_3919|M.vanbaalenii_PYR-1 AVSIWMGILLTDKLRSAFDGSP---EALAAFAEQAETPEFTGHVIDALFR
Mb3579c|M.bovis_AF2122/97 VNAVVVGMVETERSELFYGDAESIARVAATVPLGRLARPADIGWAAAFLA
Rv3549c|M.tuberculosis_H37Rv VNAVVVGMVETERSELFYGDAESIARVAATVPLGRLARPADIGWAAAFLA
MLBr_01807|M.leprae_Br4923 ANVVAPGFIDTEMTRAMD--SKYQEAALQAIPLGRIGAVEEIAGAVSFLA
MAB_0952|M.abscessus_ATCC_1997 VNAILPGNILTEGLVDMG--EEYISGMARSIPMGMLGSPVDIGHLAAFLA
. : * : * . ::
MMAR_3513|M.marinum_M DPALGELSGQTVIGAELAERYGITDEGGRRPPSHRHMLGAPRTPSTVVVR
MUL_2755|M.ulcerans_Agy99 DPALGELSGQTVIGAELAERYGITDEGGRRPPSHRHILGAPRTPSTVVVR
MAV_3035|M.avium_104 DPGLAELSGQTLIGAELAQRYGITDEGGRRPPSHRDMLGSPRTPSSVVVR
MSMEG_2923|M.smegmatis_MC2_155 DPQLGELSGQTVIGAELATRYGITDEGGRIPPSHREMLGAPRVAHPAVVR
TH_1294|M.thermoresistible__bu DPQLEQLSGHTLIGAELAERYNITDEGGRRPPSHRAMLGDPRPPSDVVVR
Mflv_2659|M.gilvum_PYR-GCK DPGLADLSGHTVIGAELAARYGITDEGGRTPPSHRGMLGEPRTPSAVVVR
Mvan_3919|M.vanbaalenii_PYR-1 DPELDDVSGHTLIAAELATRYGITDEGGRTPPSHRQMLGAPREPSPVIIR
Mb3579c|M.bovis_AF2122/97 SDAASYISGATLE-----------VHGGGEPPPYLGASSANK--------
Rv3549c|M.tuberculosis_H37Rv SDAASYISGATLE-----------VHGGGEPPPYLGASSANK--------
MLBr_01807|M.leprae_Br4923 SEDAGYISGAVIP-----------VDGGLGMGH-----------------
MAB_0952|M.abscessus_ATCC_1997 TDEAGYITGQAIV-----------VDGGQVLPESPDAVNP----------
::* .: .**