For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRALTSLDSMFLAAEDGRTVTNVSSLAVMDRTDESGKELTRADIAELLADRLHLLPPLRWRLATVPLDLG HPWWVEGEVDLDFHVRETAVVAPGDRAALETLVARLSAHPMDRSRPLWEVYLIQGLQDDKVALLTKLHHA AVDGMSGGEVLNVMFDTTPQGRVLPPAPRYRPEKEPGQLGMLARTIVGMPRRQWQSAGAARRTLTNLDQI ATLRSIPGVAAVGSAMRRAQAPIRRGPSAPSPATVTAPRLGFNGKISPHRRVALTSLPLEDVKEIKTHFE ATVNDVVVTLCAGALRRWLADRGELPEQPLVAAIPVSVRAEAEFGTYGNKVGTMLAALPTDVTDPAIRLQ RCRKELRAAKRRNEAVPASLMRDANDLVPPVLFGPAMRAMTAVAASEALSPVANLVISNVPGPRSPLYCA GRRVCEHYPVSTISDSLGLNVTVFSYTDRLEIGLVGDRYLVKDLDRLADAFGAELAVLKQSVRKPRGGRK
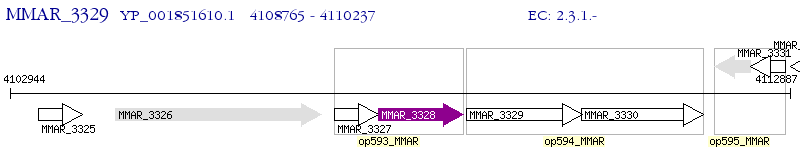
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3329 | - | - | 100% (490) | hypothetical protein MMAR_3329 |
| M. marinum M | MMAR_1605 | - | e-124 | 47.78% (496) | hypothetical protein MMAR_1605 |
| M. marinum M | MMAR_3330 | - | e-102 | 41.62% (495) | hypothetical protein MMAR_3330 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1460 | - | 2e-83 | 38.17% (482) | hypothetical protein Mb1460 |
| M. gilvum PYR-GCK | Mflv_0349 | - | 2e-80 | 38.85% (471) | hypothetical protein Mflv_0349 |
| M. tuberculosis H37Rv | Rv1425 | - | 4e-83 | 38.17% (482) | hypothetical protein Rv1425 |
| M. leprae Br4923 | MLBr_01244 | - | 2e-27 | 26.81% (429) | hypothetical protein MLBr_01244 |
| M. abscessus ATCC 19977 | MAB_0507 | - | 2e-83 | 38.30% (483) | hypothetical protein MAB_0507 |
| M. avium 104 | MAV_3352 | - | 4e-79 | 37.27% (491) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. smegmatis MC2 155 | MSMEG_4297 | - | 1e-70 | 35.23% (491) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. thermoresistible (build 8) | TH_2477 | - | 2e-69 | 36.81% (489) | PUTATIVE acyltransferase, ws/dgat/mgat subfamily protein |
| M. ulcerans Agy99 | MUL_1313 | - | 1e-101 | 41.40% (500) | hypothetical protein MUL_1313 |
| M. vanbaalenii PYR-1 | Mvan_3601 | - | 3e-71 | 34.90% (490) | hypothetical protein Mvan_3601 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3329|M.marinum_M ---------------MRALTSLDSMFLAAEDGRTVTNVSSLAVMDRTDES
MUL_1313|M.ulcerans_Agy99 ---------------MRRLSSVDAQFFAMEDGRNHSHVCQLTIVEATTTD
MSMEG_4297|M.smegmatis_MC2_155 ---------------MQRLSGLDASFLYLETAAQPMHVCSVLELDTSTMP
Mvan_3601|M.vanbaalenii_PYR-1 MDLLTPAPPRHTIGGMQRLSGLDASFLYLESAAQPLHVCSILELDTSTMP
TH_2477|M.thermoresistible__bu ------VPPPYDRG-MQRLSGLDASFLYLETPDQPLHVCSILELDTSTIP
Mb1460|M.bovis_AF2122/97 ---------------MKRLSSVDAAFWSAETAGWHMHVGALAICDPSDAP
Rv1425|M.tuberculosis_H37Rv ---------------MKRLSSVDAAFWSAETAGWHMHVGALAICDPSDAP
MAV_3352|M.avium_104 --------------MMKRLSSVDAAFWSAETAGWHMHVGALAICDPKGSA
MAB_0507|M.abscessus_ATCC_1997 ---------------MQRLSGVDAAFWHAETAGWHMHIGGLSIYDPTDAP
Mflv_0349|M.gilvum_PYR-GCK ----------------MRLSSVDAAFWFAENLKWHMHIGAIALCDPSDVP
MLBr_01244|M.leprae_Br4923 -----MADSVEGIGPFDELGALDYLLHRGEAN--PRTRAGIMAVELLDTT
* .:* : * : :
MMAR_3329|M.marinum_M GKELTRADIAELLADRLHLLPPLRWRLATVPLDLGHPWWVEG-EVDLDFH
MUL_1313|M.ulcerans_Agy99 GLRLDGQRIRGLLAQRIDLMPPLRWRVREVPFGIDYPVLVQDDTFDLDSH
MSMEG_4297|M.smegmatis_MC2_155 G-GYTFDRLRDALSLRIKAMPEFREKLADSRFNLDHPVWVEDNDFDVDRH
Mvan_3601|M.vanbaalenii_PYR-1 G-GYTFDRLRDGLALRIKAMPEFREKIADSRFNLDHPVWVEDNDFDVDRH
TH_2477|M.thermoresistible__bu G-GYDFHQLREALLLRIKAMPEFREKLADNRFNLDHPVWVEDADFDVDRH
Mb1460|M.bovis_AF2122/97 --EYSFQRLRELIIERLPEIPQLRWRVTGAPLGLDRPWFVEDEELDIDFH
Rv1425|M.tuberculosis_H37Rv --EYSFQRLRELIIERLPEIPQLRWRVTGAPLGLDRPWFVEDEELDIDFH
MAV_3352|M.avium_104 --EYTFERLRQLLIERLPEVPQLRWRVTGAPLGLDRPWFVEDEELDVDFH
MAB_0507|M.abscessus_ATCC_1997 --DYSFERVRALIVERLPSMPQLRWRVTDVPLGLDRPWFVEDEELDPDFH
Mflv_0349|M.gilvum_PYR-GCK --DFGFEQVRDLIISRLPELPQLRWRVVGAPLGLDRPYFVEDEDIDVDFH
MLBr_01244|M.leprae_Br4923 P---DWNRFRSRIEDVSQRVLRLRQKVVVPTLPTAAPRWVVDPDFELDFH
. : : :* :: : * * . .: * *
MMAR_3329|M.marinum_M VRETAVVAPGDRAALETLVARLSAHPMDRSRPLWEVYLIQGLQ------D
MUL_1313|M.ulcerans_Agy99 LWESALPAPGDDEQLGRAVADIASRPLDRARPLWELHVIHGLA------G
MSMEG_4297|M.smegmatis_MC2_155 LHRIGLPGPGGRAELAEICGHIASLPLDRSRPLWEMWVIENVAGTDAHAG
Mvan_3601|M.vanbaalenii_PYR-1 LHRIGLPSPGGRVELAEICGHIASLPLDRTRPLWEMWVIENVAGTDAHAG
TH_2477|M.thermoresistible__bu LHRIGLPAPGGPAELAEICGHLAALPLDRSRPLWEMWVIENIDGTDPRSG
Mb1460|M.bovis_AF2122/97 IRRIGVPAPGGRRELEELVGRLMSYKLDRSRPLWELWVIEGVE------G
Rv1425|M.tuberculosis_H37Rv IRRIGVPAPGGRRELEELVGRLMSYKLDRSRPLWELWVIEGVE------G
MAV_3352|M.avium_104 LRRIGVPAPGGRRELEELVGRLMSYKLDRSRPLWEMWVIEGLDGN---AG
MAB_0507|M.abscessus_ATCC_1997 IRRIGVPAPGGRKELEELAGRLMSYKLDRSRPLWELWVIEGVE------G
Mflv_0349|M.gilvum_PYR-GCK IRRIAVPSPGGRKELDDLVGRLMSYKLDRSKPLWELWFIEGLE------G
MLBr_01244|M.leprae_Br4923 VRRVRVPDPGTLREVFDLAEVIQQSPMDVSRPLWTATLVEGLA------A
: . : ** : : :* ::*** .:..:
MMAR_3329|M.marinum_M DKVALLTKLHHAAVDGMSGGEVLNVMFDTTPQ---GRVLPPAPRYRPE-K
MUL_1313|M.ulcerans_Agy99 DRVALVTKLHHSVIDGVSGMELLGALLDTVSFPDAGPELEKPPTGTAIGN
MSMEG_4297|M.smegmatis_MC2_155 GRLALMTKVHHASVDGVTGANLLSQLCTTEPD-------APPPDP-----
Mvan_3601|M.vanbaalenii_PYR-1 GRIAVMTKVHHAAVDGVTGANLMSKLCTTEPD-------APPPDP-----
TH_2477|M.thermoresistible__bu GPLVVLTKVHHAAVDGVTGANLLSQVCTTEAD-------TPPPEP-----
Mb1460|M.bovis_AF2122/97 GRIATLTKMHHAIVDGVSGAGLGEILLDITPE-------PRPPQQETVG-
Rv1425|M.tuberculosis_H37Rv GRIATLTKMHHAIVDGVSGAGLGEILLDITPE-------PRPPQQETVG-
MAV_3352|M.avium_104 GRFATLTKMHHAIVDGVSGAGLGEILLDATPE-------PRPPQQETVGS
MAB_0507|M.abscessus_ATCC_1997 GRVATLTKMHHAIVDGVSGAGLGEILLDIAPE-------PRPAPDDTVGS
Mflv_0349|M.gilvum_PYR-GCK GRVAVVTKMHHAVVDGVSGAGISEILLDTTPE-------PRPPAVDASRS
MLBr_01244|M.leprae_Br4923 GRAAMLLQISHAITDGVGSVEMFAEMYDLERDPPSRPRSPQPIPQDLSRN
. . : :: *: **: . : : .
MMAR_3329|M.marinum_M EPGQLGMLARTIVGMPRRQWQSAGAARRTLTNLDQIATLRSIPGVAAVGS
MUL_1313|M.ulcerans_Agy99 ASSDAGLLARALASLPVQPARMVRGLPPMLRHIDQLPTMRHLPGAAVIAK
MSMEG_4297|M.smegmatis_MC2_155 VDGPG----------------GGTGLEIAVNGAVRFATR-PLRLVNVVPS
Mvan_3601|M.vanbaalenii_PYR-1 VAGPG----------------GGTDLEIAVSGAVRFATR-PLKLVNVVPN
TH_2477|M.thermoresistible__bu VQGAG----------------TASDLEIALSGAVRFATR-PLKLLNVLPT
Mb1460|M.bovis_AF2122/97 FVGFQ----------------IPGLERRAIGALINVGIMTPFRIVRLLEQ
Rv1425|M.tuberculosis_H37Rv FVGFQ----------------IPGLERRAIGALINVGIMTPFRIVRLLEQ
MAV_3352|M.avium_104 LVGFQ----------------LPGLERRAVGALINVGVKTPFRIARLLEQ
MAB_0507|M.abscessus_ATCC_1997 LVGLG----------------LPSRERILLSALVNVGVKTPYRVARLVQQ
Mflv_0349|M.gilvum_PYR-GCK LVGVK----------------PPSRERQAVNGLINVWFKTPYRITRLLEQ
MLBr_01244|M.leprae_Br4923 DLMLQG---------------INHLPVALAGGVVGGLSGVASVAGRAILR
:
MMAR_3329|M.marinum_M AMRRAQAPIRRGPSAP--SPATVTAPRLGFNGKISPHRRVALTSLPLEDV
MUL_1313|M.ulcerans_Agy99 VADRAERLATRNKDGRNLQHPRAAAPKVSFGGRISAQRRFAFGSLPLDQI
MSMEG_4297|M.smegmatis_MC2_155 TLNTVVDTVRRARNGRT-MAAPFAAPRTAFNANITGHRNIAFAQLDLEDI
Mvan_3601|M.vanbaalenii_PYR-1 TVSTVIDTVKRARNGLT-MAPPFAAPKTAFNANVTGHRNVAFAQLDLEDI
TH_2477|M.thermoresistible__bu TANSVLDTVRRARAGRT-MARPFAAPSTPFNANVTGRRNVAFTQLDLEDV
Mb1460|M.bovis_AF2122/97 TVRQQIAALGVAGKP----ARYFEAPKTRFNAPVSPHRRVTGTRVELARA
Rv1425|M.tuberculosis_H37Rv TVRQQIAALGVAGKP----ARYFEAPKTRFNAPVSPHRRVTGTRVELARA
MAV_3352|M.avium_104 TVRQQVATLGVRSRP----PRYFEAPKTRFNAPVSPHRRVTGCRVELARA
MAB_0507|M.abscessus_ATCC_1997 TVRQQLAVFGMERKS----PGYFEAPHTRFNAPVSPHRRISGARLDLARV
Mflv_0349|M.gilvum_PYR-GCK TVRQQIAVRNIDNKP----PRYFDAPKVRFNGPISPHRSVAGASVPLDRV
MLBr_01244|M.leprae_Br4923 PASTVSGVVGYVRSGIR--VLSQAAEPSPLLRQRSLATRTEAIEIQLSDL
* : : : *
MMAR_3329|M.marinum_M KEIKTHFE-ATVNDVVVTLCAGALRRWLADRGELPEQPLVAAIPVSVRAE
MUL_1313|M.ulcerans_Agy99 KAAKFAVPGATVNDVVVAVCAGALRRRMIARGDDVATPLVAMVPVSVRTG
MSMEG_4297|M.smegmatis_MC2_155 KTVKNHFD-VKVNDVVMALVSGVLRTFLSHRDELPDNSLVAMVPVSVH-D
Mvan_3601|M.vanbaalenii_PYR-1 KTVKNHFG-VKVNDVVMALVSGVLRTFLLERGELPESSLVAMVPVSVH-D
TH_2477|M.thermoresistible__bu KRIKDHFG-VKVNDVVMALTSGALRSYLLERDQLPDSPLIAMVPVSVH-D
Mb1460|M.bovis_AF2122/97 KAVKDAFG-VKLNDVVLALVAGAARQYLQKRDELPAKPLIAQIPVSTRSE
Rv1425|M.tuberculosis_H37Rv KAVKDAFG-VKLNDVVLALVAGAARQYLQKRDELPAKPLIAQIPVSTRSE
MAV_3352|M.avium_104 KAIKDAYD-VKLNDVVLALVAGAVRDYLQKRGELPAKPLIAQIPVSTRTD
MAB_0507|M.abscessus_ATCC_1997 KALKDAFG-VKVNDVVLAIVAGAVRDYLAKRDEFPDKPLIVQIPVSVRTE
Mflv_0349|M.gilvum_PYR-GCK KAVKNAFD-VKLNDVVLALVSGALRSYLKDRGELPAKSLVSQVPVSTR-A
MLBr_01244|M.leprae_Br4923 HKAAKAGD-GSINDAYLAGLVGALRRYHEALG-VSISTLPMAVPVNVR-T
: .:**. :: *. * . .* :**..:
MMAR_3329|M.marinum_M AEFGTYGNKVGTMLAALPTDVTDPAIRLQRCRKELRAAKRRNEAVPASLM
MUL_1313|M.ulcerans_Agy99 ERQ--YGNQISSMIVPIPTDQADARARLISAHQVMRAAKERHRAIPAKLL
MSMEG_4297|M.smegmatis_MC2_155 RSDRPGRNQVSGMFSPLQTHIADPAERLRAIAEANAVAKQHSSAIGATLL
Mvan_3601|M.vanbaalenii_PYR-1 KSDRPGRNQVSGMFSRLETNIEDPATRLKAIAEANSVAKQHSSAIAATLL
TH_2477|M.thermoresistible__bu KTDRPGRNHVSGMFSRLETTIEDPAERLRAIANANSVAKQHSSAIAASLL
Mb1460|M.bovis_AF2122/97 ETKADVGNQVSSMTASLATHIEDPAKRLAAIHESTLSAKEMAKALSAHQI
Rv1425|M.tuberculosis_H37Rv ETKADVGNQVSSMTASLATHIEDPAKRLAAIHESTLSAKEMAKAPSAHQI
MAV_3352|M.avium_104 ENKDDIGNKVSSMTVSLATHIEDPGERLRAIHDSTQSAKLMAKALSAHQI
MAB_0507|M.abscessus_ATCC_1997 ETDGVVGNQVSSMTVSLATDIDDPAERIQAIYEGTQGAKEMARAMSAHQI
Mflv_0349|M.gilvum_PYR-GCK QADDTVGNQVNAMTVVLATDVADPAERIKAIYSYTQGAKEMTKALTAHQI
MLBr_01244|M.leprae_Br4923 EADVVGSNRFVGVTLAAPLGTNDPAARMQKIRSQMTQRRDEPAMNIIGSL
*:. : *. *: . : :
MMAR_3329|M.marinum_M RDANDLVPPVLFGPAMRAMTAVAASEALSPVANLVISNVPGPRSPLYCAG
MUL_1313|M.ulcerans_Agy99 TEANHTVPPALLTRAARVISMTTGSGWIRPPFNVTISNIPGSPDPLYCAG
MSMEG_4297|M.smegmatis_MC2_155 QDWTQFAAPAVFGTAMRVYAASRLSGAK-PVHNLVVSNVPGPQVPLYFLG
Mvan_3601|M.vanbaalenii_PYR-1 QDWTQFAAPAVFGAAMRVYASSRLSGAR-PVHNLVISNVPGPQGPLYMLG
TH_2477|M.thermoresistible__bu QDWTQFAAPAVFGVAMRVYAASRLGEAK-PVHNLVVSNVPGPQVPLYFLG
Mb1460|M.bovis_AF2122/97 MGLTETTPPGLLQLAARAYTASGLSHNL-APINLVVSNVPGPPFPLYMAG
Rv1425|M.tuberculosis_H37Rv MGLTETTPPGLLQLAARAYTASGLSHNL-APINLVVSNVPGPPFPLYMAG
MAV_3352|M.avium_104 MGLTETTPPGLLHLAARAYTASGLSRNL-APINLVVSNVPGPPFPLYMAG
MAB_0507|M.abscessus_ATCC_1997 MGLTDTTPPGLLQLAARAYTASGLSRNL-APINLVLSNVPGPSFPLYMAG
Mflv_0349|M.gilvum_PYR-GCK MGYTETTPPGLLALASRAYAASGIGSNI-APMNVVISNVPGPTFPLYLGG
MLBr_01244|M.leprae_Br4923 APLMTVLPASVLDFIVDSVASS----------DVNASNIPAYPGDTYFAG
.. :: : :: **:*. * *
MMAR_3329|M.marinum_M RRVCEHYPVSTISDSLGLNVTVFSYTDRLEIGLVGDRYLVKDLDRLADAF
MUL_1313|M.ulcerans_Agy99 ARVVSQHPVNVLLDGMGLSITLLSYQDRLDFGLTADRELLPDVWHLADEL
MSMEG_4297|M.smegmatis_MC2_155 AEVKAMYPLGPIFHGSGLNITVMSLTGKLDVGIISCPELLPDLWDMADDF
Mvan_3601|M.vanbaalenii_PYR-1 CEVKAMYPLGPIFHGSGLNMTVMSLSGKLDVGIVSCPELLPDLWDMADTF
TH_2477|M.thermoresistible__bu AEVKAMYPLGPIFHGSGLNITVMSLTGRLNVGMVSCPELMPDLWELAEGF
Mb1460|M.bovis_AF2122/97 ARLDSLVPLGPPVMDVALNITCFSYQDYLDFGLVTTPEVANDIDEMADAI
Rv1425|M.tuberculosis_H37Rv ARLDSLVPLGPPVMDVALNITCFSYQDYLDFGLVTTPEVANDIDEMADAI
MAV_3352|M.avium_104 AKLDSLVPLGPPVMDVALNITCFSYTDYLDFGFVTTPEVANDIDDMADRI
MAB_0507|M.abscessus_ATCC_1997 ARLESMIPLGPPVMDVALNITCFSYLEHMDFGFVTTPEVAADIDEMADGL
Mflv_0349|M.gilvum_PYR-GCK ARVEHLMPVGPLLFDVALNVTCFSYCGAVDFGFITTPEIAADIVDLADQI
MLBr_01244|M.leprae_Br4923 AKILRQYGIGP-RPGVAMMAVLMSRGGFCTVTVRYDRASVKSEALFARCL
.: :. . .: . :* . . . :* :
MMAR_3329|M.marinum_M GAELAVLKQSVRKPRG-----------GRK---------
MUL_1313|M.ulcerans_Agy99 AAELSALAAEVGAPRNPVADGAPTSTTGRPTAKPGAIPR
MSMEG_4297|M.smegmatis_MC2_155 AVALDELLDATKRA-------------------------
Mvan_3601|M.vanbaalenii_PYR-1 GVALQELLDATR---------------------------
TH_2477|M.thermoresistible__bu ERALRELLDTVG---------------------------
Mb1460|M.bovis_AF2122/97 EPALAELERAAE---------------------------
Rv1425|M.tuberculosis_H37Rv EPALAELERAAE---------------------------
MAV_3352|M.avium_104 EPALTDLEKAAAGETG-----------------------
MAB_0507|M.abscessus_ATCC_1997 EAALVELEHAVGI--------------------------
Mflv_0349|M.gilvum_PYR-GCK EPALHDLEVAAGLVSDPTRPKPRKRGRR-----------
MLBr_01244|M.leprae_Br4923 LEGFDEVLALAGDPTPHAVPASFAARSSGSPAGWLSSS-
: : .