For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRRLSSVDAQFFAMEDGRNHSHVCQLTIVEATTTDGLRLDGQRIRGLLAQRIDLMPPLRWRVREVPFGID YPVLVQDDTFDLDSHLWESALPAPGDDEQLGRAVADIASRPLDRARPLWELHVIHGLAGDRVALVTKLHH SVIDGVSGMELLGALLDTVPFPDAGPELEKPPTGTAIGNAPSDAGLLARALASLPVQPARMVRGLPPMLR HIDQLPTMRHLPGAAVIAKVADRAERLATRNKDGRNLQHPRAAAPKVSFGGRISAQRRFAFGSLPLDQIK AAKFAVPGATVNDVVVAVCAGALRRRMIARGDDVATPLVAMVPVSVRTGERQYGNQISSMIVPIPTDQAD ARTRLISAHQVMRAAKERHRAIPAKLLTEANHTVPPALLTRAARVISMTTGSGWIRPPFNVTISNIPGSP DPLYCAGARVVSQHPVNVLLDGMGLSITLLSYQDRLDFGLTADRELLPDVWHLADELAAELSALAAEVGA PRNPVADGAPTGTTGRPTAKPGAIPR
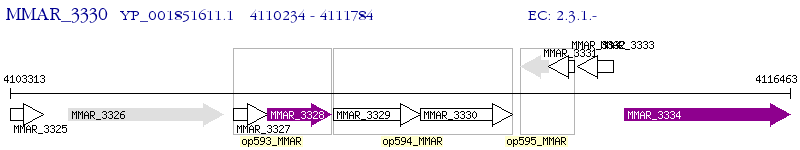
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3330 | - | - | 100% (516) | hypothetical protein MMAR_3330 |
| M. marinum M | MMAR_1605 | - | e-134 | 50.51% (491) | hypothetical protein MMAR_1605 |
| M. marinum M | MMAR_3329 | - | e-102 | 41.62% (495) | hypothetical protein MMAR_3329 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1460 | - | 5e-73 | 37.07% (491) | hypothetical protein Mb1460 |
| M. gilvum PYR-GCK | Mflv_0349 | - | 4e-72 | 35.96% (495) | hypothetical protein Mflv_0349 |
| M. tuberculosis H37Rv | Rv1425 | - | 2e-72 | 37.07% (491) | hypothetical protein Rv1425 |
| M. leprae Br4923 | MLBr_01244 | - | 3e-21 | 27.32% (366) | hypothetical protein MLBr_01244 |
| M. abscessus ATCC 19977 | MAB_0507 | - | 2e-73 | 36.97% (495) | hypothetical protein MAB_0507 |
| M. avium 104 | MAV_2240 | - | 5e-70 | 35.70% (493) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. smegmatis MC2 155 | MSMEG_4297 | - | 3e-71 | 35.85% (491) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. thermoresistible (build 8) | TH_2477 | - | 1e-64 | 33.00% (497) | PUTATIVE acyltransferase, ws/dgat/mgat subfamily protein |
| M. ulcerans Agy99 | MUL_1313 | - | 0.0 | 99.22% (516) | hypothetical protein MUL_1313 |
| M. vanbaalenii PYR-1 | Mvan_3601 | - | 4e-71 | 35.03% (491) | hypothetical protein Mvan_3601 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3330|M.marinum_M ---------------MRRLSSVDAQFFAMEDGRNHSHVCQLTIVEATTTD
MUL_1313|M.ulcerans_Agy99 ---------------MRRLSSVDAQFFAMEDGRNHSHVCQLTIVEATTTD
MSMEG_4297|M.smegmatis_MC2_155 ---------------MQRLSGLDASFLYLETAAQPMHVCSVLELDTSTMP
Mvan_3601|M.vanbaalenii_PYR-1 MDLLTPAPPRHTIGGMQRLSGLDASFLYLESAAQPLHVCSILELDTSTMP
TH_2477|M.thermoresistible__bu ------VPPPYDRG-MQRLSGLDASFLYLETPDQPLHVCSILELDTSTIP
MAV_2240|M.avium_104 ------------MLRMQRLSGLDASFLYLETSSQPMHVCSIIELDTSTVP
Mb1460|M.bovis_AF2122/97 ---------------MKRLSSVDAAFWSAETAGWHMHVGALAICDPSDAP
Rv1425|M.tuberculosis_H37Rv ---------------MKRLSSVDAAFWSAETAGWHMHVGALAICDPSDAP
MAB_0507|M.abscessus_ATCC_1997 ---------------MQRLSGVDAAFWHAETAGWHMHIGGLSIYDPTDAP
Mflv_0349|M.gilvum_PYR-GCK ----------------MRLSSVDAAFWFAENLKWHMHIGAIALCDPSDVP
MLBr_01244|M.leprae_Br4923 -----MADSVEGIGPFDELGALDYLLHRGEAN--PRTRAGIMAVELLDTT
.*..:* : * : :
MMAR_3330|M.marinum_M GLRLDGQRIRGLLAQRIDLMPPLRWRVREVPFGIDYPVLVQDDTFDLDSH
MUL_1313|M.ulcerans_Agy99 GLRLDGQRIRGLLAQRIDLMPPLRWRVREVPFGIDYPVLVQDDTFDLDSH
MSMEG_4297|M.smegmatis_MC2_155 G-GYTFDRLRDALSLRIKAMPEFREKLADSRFNLDHPVWVEDNDFDVDRH
Mvan_3601|M.vanbaalenii_PYR-1 G-GYTFDRLRDGLALRIKAMPEFREKIADSRFNLDHPVWVEDNDFDVDRH
TH_2477|M.thermoresistible__bu G-GYDFHQLREALLLRIKAMPEFREKLADNRFNLDHPVWVEDADFDVDRH
MAV_2240|M.avium_104 G-GYTFERLRDALVRRIRAVPPFREKLADSPLNLDHPVWVDDEDFDVDRH
Mb1460|M.bovis_AF2122/97 --EYSFQRLRELIIERLPEIPQLRWRVTGAPLGLDRPWFVEDEELDIDFH
Rv1425|M.tuberculosis_H37Rv --EYSFQRLRELIIERLPEIPQLRWRVTGAPLGLDRPWFVEDEELDIDFH
MAB_0507|M.abscessus_ATCC_1997 --DYSFERVRALIVERLPSMPQLRWRVTDVPLGLDRPWFVEDEELDPDFH
Mflv_0349|M.gilvum_PYR-GCK --DFGFEQVRDLIISRLPELPQLRWRVVGAPLGLDRPYFVEDEDIDVDFH
MLBr_01244|M.leprae_Br4923 P---DWNRFRSRIEDVSQRVLRLRQKVVVPTLPTAAPRWVVDPDFELDFH
.:.* : : :* :: : * * * :: * *
MMAR_3330|M.marinum_M LWESALPAPGDDEQLGRAVADIASRPLDRARPLWELHVIHGLA------G
MUL_1313|M.ulcerans_Agy99 LWESALPAPGDDEQLGRAVADIASRPLDRARPLWELHVIHGLA------G
MSMEG_4297|M.smegmatis_MC2_155 LHRIGLPGPGGRAELAEICGHIASLPLDRSRPLWEMWVIENVAGTDAHAG
Mvan_3601|M.vanbaalenii_PYR-1 LHRIGLPSPGGRVELAEICGHIASLPLDRTRPLWEMWVIENVAGTDAHAG
TH_2477|M.thermoresistible__bu LHRIGLPAPGGPAELAEICGHLAALPLDRSRPLWEMWVIENIDGTDPRSG
MAV_2240|M.avium_104 LHRIAVPAPGGRAELSQICGHIAQTPLDRRRPLWEMWVIEGVAGTDCHRD
Mb1460|M.bovis_AF2122/97 IRRIGVPAPGGRRELEELVGRLMSYKLDRSRPLWELWVIEGVE------G
Rv1425|M.tuberculosis_H37Rv IRRIGVPAPGGRRELEELVGRLMSYKLDRSRPLWELWVIEGVE------G
MAB_0507|M.abscessus_ATCC_1997 IRRIGVPAPGGRKELEELAGRLMSYKLDRSRPLWELWVIEGVE------G
Mflv_0349|M.gilvum_PYR-GCK IRRIAVPSPGGRKELDDLVGRLMSYKLDRSKPLWELWFIEGLE------G
MLBr_01244|M.leprae_Br4923 VRRVRVPDPGTLREVFDLAEVIQQSPMDVSRPLWTATLVEGLA------A
: . :* ** :: : :* :*** .:..:
MMAR_3330|M.marinum_M DRVALVTKLHHSVIDGVSGMELLGALLDTVPFPDAGPELEK------PPT
MUL_1313|M.ulcerans_Agy99 DRVALVTKLHHSVIDGVSGMELLGALLDTVSFPDAGPELEK------PPT
MSMEG_4297|M.smegmatis_MC2_155 GRLALMTKVHHASVDGVTGANLLSQLCTTEPDAPP-PDP-----------
Mvan_3601|M.vanbaalenii_PYR-1 GRIAVMTKVHHAAVDGVTGANLMSKLCTTEPDAPP-PDP-----------
TH_2477|M.thermoresistible__bu GPLVVLTKVHHAAVDGVTGANLLSQVCTTEADTPP-PEP-----------
MAV_2240|M.avium_104 GRLAVMTKVHHAGVDGVTGANLMSQLCATEPDAAP-PEP-----------
Mb1460|M.bovis_AF2122/97 GRIATLTKMHHAIVDGVSGAGLGEILLDITPEPRP-PQQET------VG-
Rv1425|M.tuberculosis_H37Rv GRIATLTKMHHAIVDGVSGAGLGEILLDITPEPRP-PQQET------VG-
MAB_0507|M.abscessus_ATCC_1997 GRVATLTKMHHAIVDGVSGAGLGEILLDIAPEPRP-APDDT------VGS
Mflv_0349|M.gilvum_PYR-GCK GRVAVVTKMHHAVVDGVSGAGISEILLDTTPEPRP-PAVDA------SRS
MLBr_01244|M.leprae_Br4923 GRAAMLLQISHAITDGVGSVEMFAEMYDLERDPPSRPRSPQPIPQDLSRN
. . : :: *: *** . : : . . .
MMAR_3330|M.marinum_M GTAIGNAPSDAGLLARALASLPVQPARMVRGLPPMLRHIDQLPTMRHLPG
MUL_1313|M.ulcerans_Agy99 GTAIGNASSDAGLLARALASLPVQPARMVRGLPPMLRHIDQLPTMRHLPG
MSMEG_4297|M.smegmatis_MC2_155 VDGPGGGTGLEIAVNGAVRFATR-PLRLVN--------------------
Mvan_3601|M.vanbaalenii_PYR-1 VAGPGGGTDLEIAVSGAVRFATR-PLKLVN--------------------
TH_2477|M.thermoresistible__bu VQGAGTASDLEIALSGAVRFATR-PLKLLN--------------------
MAV_2240|M.avium_104 AAGVGGGAGWRIAAGGLVRFAAR-PLRLAS--------------------
Mb1460|M.bovis_AF2122/97 FVGFQIPGLERRAIGALINVGIMTPFRIVR--------------------
Rv1425|M.tuberculosis_H37Rv FVGFQIPGLERRAIGALINVGIMTPFRIVR--------------------
MAB_0507|M.abscessus_ATCC_1997 LVGLGLPSRERILLSALVNVGVKTPYRVAR--------------------
Mflv_0349|M.gilvum_PYR-GCK LVGVKPPSRERQAVNGLINVWFKTPYRITR--------------------
MLBr_01244|M.leprae_Br4923 DLMLQGINHLPVALAGGVVGGLSGVASVAG--------------------
: :
MMAR_3330|M.marinum_M AAVIAKVADRAERLATRNKDGRNLQHPRAAAPKVSFGGRISAQRRFAFGS
MUL_1313|M.ulcerans_Agy99 AAVIAKVADRAERLATRNKDGRNLQHPRAAAPKVSFGGRISAQRRFAFGS
MSMEG_4297|M.smegmatis_MC2_155 --VVPSTLNTVVDTVRRARNGR-TMAAPFAAPRTAFNANITGHRNIAFAQ
Mvan_3601|M.vanbaalenii_PYR-1 --VVPNTVSTVIDTVKRARNGL-TMAPPFAAPKTAFNANVTGHRNVAFAQ
TH_2477|M.thermoresistible__bu --VLPTTANSVLDTVRRARAGR-TMARPFAAPSTPFNANVTGRRNVAFTQ
MAV_2240|M.avium_104 --VVPDTAASVLATVRRALDGA-AMARPFAAPTTVFNARISNRRCIAYAE
Mb1460|M.bovis_AF2122/97 --LLEQTVRQQIAALGVAGKP----ARYFEAPKTRFNAPVSPHRRVTGTR
Rv1425|M.tuberculosis_H37Rv --LLEQTVRQQIAALGVAGKP----ARYFEAPKTRFNAPVSPHRRVTGTR
MAB_0507|M.abscessus_ATCC_1997 --LVQQTVRQQLAVFGMERKS----PGYFEAPHTRFNAPVSPHRRISGAR
Mflv_0349|M.gilvum_PYR-GCK --LLEQTVRQQIAVRNIDNKP----PRYFDAPKVRFNGPISPHRSVAGAS
MLBr_01244|M.leprae_Br4923 -RAILRPASTVSGVVGYVRSG--IRVLSQAAEPSPLLRQRSLATRTEAIE
: * : :
MMAR_3330|M.marinum_M LPLDQIKAAKFAVPGATVNDVVVAVCAGALRRRMIARGDDVATPLVAMVP
MUL_1313|M.ulcerans_Agy99 LPLDQIKAAKFAVPGATVNDVVVAVCAGALRRRMIARGDDVATPLVAMVP
MSMEG_4297|M.smegmatis_MC2_155 LDLEDIKTVKNHF-DVKVNDVVMALVSGVLRTFLSHRDELPDNSLVAMVP
Mvan_3601|M.vanbaalenii_PYR-1 LDLEDIKTVKNHF-GVKVNDVVMALVSGVLRTFLLERGELPESSLVAMVP
TH_2477|M.thermoresistible__bu LDLEDVKRIKDHF-GVKVNDVVMALTSGALRSYLLERDQLPDSPLIAMVP
MAV_2240|M.avium_104 LNLDDVKAVKNHF-GVKVNDVVMALVSGVLRQYLAERNALPDSSLVASVP
Mb1460|M.bovis_AF2122/97 VELARAKAVKDAF-GVKLNDVVLALVAGAARQYLQKRDELPAKPLIAQIP
Rv1425|M.tuberculosis_H37Rv VELARAKAVKDAF-GVKLNDVVLALVAGAARQYLQKRDELPAKPLIAQIP
MAB_0507|M.abscessus_ATCC_1997 LDLARVKALKDAF-GVKVNDVVLAIVAGAVRDYLAKRDEFPDKPLIVQIP
Mflv_0349|M.gilvum_PYR-GCK VPLDRVKAVKNAF-DVKLNDVVLALVSGALRSYLKDRGELPAKSLVSQVP
MLBr_01244|M.leprae_Br4923 IQLSDLHKAAKAG-DGSINDAYLAGLVGALRRYHEALG-VSISTLPMAVP
: * : . .:**. :* *. * . ..* :*
MMAR_3330|M.marinum_M VSVR--TGERQYGNQISSMIVPIPTDQADARTRLISAHQVMRAAKERHRA
MUL_1313|M.ulcerans_Agy99 VSVR--TGERQYGNQISSMIVPIPTDQADARARLISAHQVMRAAKERHRA
MSMEG_4297|M.smegmatis_MC2_155 VSVH-DRSDRPGRNQVSGMFSPLQTHIADPAERLRAIAEANAVAKQHSSA
Mvan_3601|M.vanbaalenii_PYR-1 VSVH-DKSDRPGRNQVSGMFSRLETNIEDPATRLKAIAEANSVAKQHSSA
TH_2477|M.thermoresistible__bu VSVH-DKTDRPGRNHVSGMFSRLETTIEDPAERLRAIANANSVAKQHSSA
MAV_2240|M.avium_104 ISVH-GKSDRPGRNQVSAMFASLHTEIADPVQRLKAIAHANSLAKEHSSA
Mb1460|M.bovis_AF2122/97 VSTRSEETKADVGNQVSSMTASLATHIEDPAKRLAAIHESTLSAKEMAKA
Rv1425|M.tuberculosis_H37Rv VSTRSEETKADVGNQVSSMTASLATHIEDPAKRLAAIHESTLSAKEMAKA
MAB_0507|M.abscessus_ATCC_1997 VSVRTEETDGVVGNQVSSMTVSLATDIDDPAERIQAIYEGTQGAKEMARA
Mflv_0349|M.gilvum_PYR-GCK VSTR-AQADDTVGNQVNAMTVVLATDVADPAERIKAIYSYTQGAKEMTKA
MLBr_01244|M.leprae_Br4923 VNVR-TEADVVGSNRFVGVTLAAPLGTNDPAARMQKIRSQMTQRRDEPAM
:..: . *:. .: *. *: ::
MMAR_3330|M.marinum_M IPAKLLTEANHTVPPALLTRAARVISMTTGSGWIRPPFNVTISNIPGSPD
MUL_1313|M.ulcerans_Agy99 IPAKLLTEANHTVPPALLTRAARVISMTTGSGWIRPPFNVTISNIPGSPD
MSMEG_4297|M.smegmatis_MC2_155 IGATLLQDWTQFAAPAVFGTAMRVYAASR-LSGAKPVHNLVVSNVPGPQV
Mvan_3601|M.vanbaalenii_PYR-1 IAATLLQDWTQFAAPAVFGAAMRVYASSR-LSGARPVHNLVISNVPGPQG
TH_2477|M.thermoresistible__bu IAASLLQDWTQFAAPAVFGVAMRVYAASR-LGEAKPVHNLVVSNVPGPQV
MAV_2240|M.avium_104 IGASLLQDWTQFAAPAVFGIAMRLYARTR-FTDSMPVHNLVVSNVPGPQL
Mb1460|M.bovis_AF2122/97 LSAHQIMGLTETTPPGLLQLAARAYTASG-LSHNLAPINLVVSNVPGPPF
Rv1425|M.tuberculosis_H37Rv PSAHQIMGLTETTPPGLLQLAARAYTASG-LSHNLAPINLVVSNVPGPPF
MAB_0507|M.abscessus_ATCC_1997 MSAHQIMGLTDTTPPGLLQLAARAYTASG-LSRNLAPINLVLSNVPGPSF
Mflv_0349|M.gilvum_PYR-GCK LTAHQIMGYTETTPPGLLALASRAYAASG-IGSNIAPMNVVISNVPGPTF
MLBr_01244|M.leprae_Br4923 NIIGSLAPLMTVLPASVLDFIVDSVASSD----------VNASNIPAYPG
: ...:: : : : **:*.
MMAR_3330|M.marinum_M PLYCAGARVVSQHPVNVLLDGMGLSITLLSYQDRLDFGLTADRELLPDVW
MUL_1313|M.ulcerans_Agy99 PLYCAGARVVSQHPVNVLLDGMGLSITLLSYQDRLDFGLTADRELLPDVW
MSMEG_4297|M.smegmatis_MC2_155 PLYFLGAEVKAMYPLGPIFHGSGLNITVMSLTGKLDVGIISCPELLPDLW
Mvan_3601|M.vanbaalenii_PYR-1 PLYMLGCEVKAMYPLGPIFHGSGLNMTVMSLSGKLDVGIVSCPELLPDLW
TH_2477|M.thermoresistible__bu PLYFLGAEVKAMYPLGPIFHGSGLNITVMSLTGRLNVGMVSCPELMPDLW
MAV_2240|M.avium_104 PLYLLGCEVKAMYPLGPIFHGSGLNITAMSLRGKLDVGLIGCPDLVPDLW
Mb1460|M.bovis_AF2122/97 PLYMAGARLDSLVPLGPPVMDVALNITCFSYQDYLDFGLVTTPEVANDID
Rv1425|M.tuberculosis_H37Rv PLYMAGARLDSLVPLGPPVMDVALNITCFSYQDYLDFGLVTTPEVANDID
MAB_0507|M.abscessus_ATCC_1997 PLYMAGARLESMIPLGPPVMDVALNITCFSYLEHMDFGFVTTPEVAADID
Mflv_0349|M.gilvum_PYR-GCK PLYLGGARVEHLMPVGPLLFDVALNVTCFSYCGAVDFGFITTPEIAADIV
MLBr_01244|M.leprae_Br4923 DTYFAGAKILRQYGIGP-RPGVAMMAVLMSRGGFCTVTVRYDRASVKSEA
* *..: :. . .: . :* . . .
MMAR_3330|M.marinum_M HLADELAAELSALAAEVGAPRNPVADGAPTGTTGRPTAKPGAIPR
MUL_1313|M.ulcerans_Agy99 HLADELAAELSALAAEVGAPRNPVADGAPTSTTGRPTAKPGAIPR
MSMEG_4297|M.smegmatis_MC2_155 DMADDFAVALDELLDATKRA-------------------------
Mvan_3601|M.vanbaalenii_PYR-1 DMADTFGVALQELLDATR---------------------------
TH_2477|M.thermoresistible__bu ELAEGFERALRELLDTVG---------------------------
MAV_2240|M.avium_104 ELADEFAVAMEELRAAAR---------------------------
Mb1460|M.bovis_AF2122/97 EMADAIEPALAELERAAE---------------------------
Rv1425|M.tuberculosis_H37Rv EMADAIEPALAELERAAE---------------------------
MAB_0507|M.abscessus_ATCC_1997 EMADGLEAALVELEHAVGI--------------------------
Mflv_0349|M.gilvum_PYR-GCK DLADQIEPALHDLEVAAGLVSDPTRPKPRKRGRR-----------
MLBr_01244|M.leprae_Br4923 LFARCLLEGFDEVLALAGDPTPHAVPASFAARSSGSPAGWLSSS-
:* : : : .