For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHQLTSLDAQFLAAEDGRTHGHVSALGIYDPSTAPGARLTLEAVRDLVATRIDSLAPFRWRLAPVPLNLD HPYWFDDPVFDLEFHIRELALPAPGDDRQLAEQVARLHSRPLDRGRPLWELYLIQGLSGGRVAIMTKIHH AAVDGVSGGELLSVLLDPDPNPRTLERLEPAQRPPRRGDPVPGQVGLLLRGLLGLARQPLRALAATPRTL PHLDQVPTVRMLPGVRALAAGSALLTGARRRDGGVLERTNLRAPRTRFNRRIGPHRRFAFASVPLADVKA VKNAMGVTVNDVVLALSAGVLRRRLAAAGDLPTEPLLAMVPVSVRTPEQAGTFGNRVSTMIVPLPTDEPD ARRRVARVNDVMRSAKQRHRAIPATLMQDANQFIPPALLARASRVVGQLSVSDPLAPPVNVVVSNVPGSP TPLYLAGARLQAQFPVSIVIDGVGVNLTVLSYRDSLDIGVIADRDLVEDAWPFIEDIKAELAELSALVIP RPKRKTRTRTARTQPKEDPK
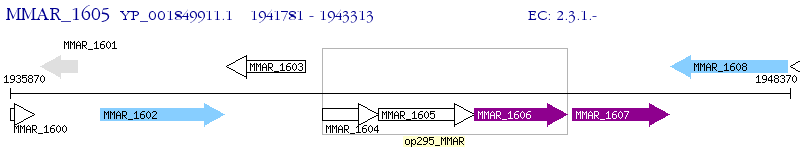
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1605 | - | - | 100% (510) | hypothetical protein MMAR_1605 |
| M. marinum M | MMAR_3330 | - | e-134 | 50.51% (491) | hypothetical protein MMAR_3330 |
| M. marinum M | MMAR_3329 | - | e-124 | 47.78% (496) | hypothetical protein MMAR_3329 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1460 | - | 4e-89 | 40.49% (489) | hypothetical protein Mb1460 |
| M. gilvum PYR-GCK | Mflv_0349 | - | 3e-84 | 38.22% (505) | hypothetical protein Mflv_0349 |
| M. tuberculosis H37Rv | Rv1425 | - | 1e-88 | 40.49% (489) | hypothetical protein Rv1425 |
| M. leprae Br4923 | MLBr_01244 | - | 4e-27 | 30.07% (429) | hypothetical protein MLBr_01244 |
| M. abscessus ATCC 19977 | MAB_0507 | - | 5e-90 | 40.37% (488) | hypothetical protein MAB_0507 |
| M. avium 104 | MAV_2240 | - | 6e-86 | 39.60% (495) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. smegmatis MC2 155 | MSMEG_4297 | - | 4e-84 | 40.04% (497) | acyltransferase, ws/dgat/mgat subfamily protein |
| M. thermoresistible (build 8) | TH_2477 | - | 8e-76 | 36.99% (492) | PUTATIVE acyltransferase, ws/dgat/mgat subfamily protein |
| M. ulcerans Agy99 | MUL_1313 | - | 1e-132 | 50.10% (491) | hypothetical protein MUL_1313 |
| M. vanbaalenii PYR-1 | Mvan_3601 | - | 1e-81 | 37.30% (496) | hypothetical protein Mvan_3601 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1605|M.marinum_M ---------------MHQLTSLDAQFLAAEDGRTHGHVSALGIYDPSTAP
MUL_1313|M.ulcerans_Agy99 ---------------MRRLSSVDAQFFAMEDGRNHSHVCQLTIVEATTTD
MSMEG_4297|M.smegmatis_MC2_155 ---------------MQRLSGLDASFLYLETAAQPMHVCSVLELDTSTMP
Mvan_3601|M.vanbaalenii_PYR-1 MDLLTPAPPRHTIGGMQRLSGLDASFLYLESAAQPLHVCSILELDTSTMP
TH_2477|M.thermoresistible__bu ------VPPPYDRG-MQRLSGLDASFLYLETPDQPLHVCSILELDTSTIP
MAV_2240|M.avium_104 ------------MLRMQRLSGLDASFLYLETSSQPMHVCSIIELDTSTVP
Mb1460|M.bovis_AF2122/97 ---------------MKRLSSVDAAFWSAETAGWHMHVGALAICDPSDAP
Rv1425|M.tuberculosis_H37Rv ---------------MKRLSSVDAAFWSAETAGWHMHVGALAICDPSDAP
MAB_0507|M.abscessus_ATCC_1997 ---------------MQRLSGVDAAFWHAETAGWHMHIGGLSIYDPTDAP
Mflv_0349|M.gilvum_PYR-GCK ----------------MRLSSVDAAFWFAENLKWHMHIGAIALCDPSDVP
MLBr_01244|M.leprae_Br4923 -----MADSVEGIGPFDELGALDYLLHRGEAN--PRTRAGIMAVELLDTT
.* .:* : * : :
MMAR_1605|M.marinum_M GARLTLEAVRDLVATRIDSLAPFRWRLAPVPLNLDHPYWFDDPVFDLEFH
MUL_1313|M.ulcerans_Agy99 GLRLDGQRIRGLLAQRIDLMPPLRWRVREVPFGIDYPVLVQDDTFDLDSH
MSMEG_4297|M.smegmatis_MC2_155 G-GYTFDRLRDALSLRIKAMPEFREKLADSRFNLDHPVWVEDNDFDVDRH
Mvan_3601|M.vanbaalenii_PYR-1 G-GYTFDRLRDGLALRIKAMPEFREKIADSRFNLDHPVWVEDNDFDVDRH
TH_2477|M.thermoresistible__bu G-GYDFHQLREALLLRIKAMPEFREKLADNRFNLDHPVWVEDADFDVDRH
MAV_2240|M.avium_104 G-GYTFERLRDALVRRIRAVPPFREKLADSPLNLDHPVWVDDEDFDVDRH
Mb1460|M.bovis_AF2122/97 --EYSFQRLRELIIERLPEIPQLRWRVTGAPLGLDRPWFVEDEELDIDFH
Rv1425|M.tuberculosis_H37Rv --EYSFQRLRELIIERLPEIPQLRWRVTGAPLGLDRPWFVEDEELDIDFH
MAB_0507|M.abscessus_ATCC_1997 --DYSFERVRALIVERLPSMPQLRWRVTDVPLGLDRPWFVEDEELDPDFH
Mflv_0349|M.gilvum_PYR-GCK --DFGFEQVRDLIISRLPELPQLRWRVVGAPLGLDRPYFVEDEDIDVDFH
MLBr_01244|M.leprae_Br4923 P---DWNRFRSRIEDVSQRVLRLRQKVVVPTLPTAAPRWVVDPDFELDFH
. .* : : :* :: : * . * :: : *
MMAR_1605|M.marinum_M IRELALPAPGDDRQLAEQVARLHSRPLDRGRPLWELYLIQGLS------G
MUL_1313|M.ulcerans_Agy99 LWESALPAPGDDEQLGRAVADIASRPLDRARPLWELHVIHGLA------G
MSMEG_4297|M.smegmatis_MC2_155 LHRIGLPGPGGRAELAEICGHIASLPLDRSRPLWEMWVIENVAGTDAHAG
Mvan_3601|M.vanbaalenii_PYR-1 LHRIGLPSPGGRVELAEICGHIASLPLDRTRPLWEMWVIENVAGTDAHAG
TH_2477|M.thermoresistible__bu LHRIGLPAPGGPAELAEICGHLAALPLDRSRPLWEMWVIENIDGTDPRSG
MAV_2240|M.avium_104 LHRIAVPAPGGRAELSQICGHIAQTPLDRRRPLWEMWVIEGVAGTDCHRD
Mb1460|M.bovis_AF2122/97 IRRIGVPAPGGRRELEELVGRLMSYKLDRSRPLWELWVIEGVE------G
Rv1425|M.tuberculosis_H37Rv IRRIGVPAPGGRRELEELVGRLMSYKLDRSRPLWELWVIEGVE------G
MAB_0507|M.abscessus_ATCC_1997 IRRIGVPAPGGRKELEELAGRLMSYKLDRSRPLWELWVIEGVE------G
Mflv_0349|M.gilvum_PYR-GCK IRRIAVPSPGGRKELDDLVGRLMSYKLDRSKPLWELWFIEGLE------G
MLBr_01244|M.leprae_Br4923 VRRVRVPDPGTLREVFDLAEVIQQSPMDVSRPLWTATLVEGLA------A
: . :* ** :: : :* :*** .:..:
MMAR_1605|M.marinum_M GRVAIMTKIHHAAVDGVSGGELLSVLLDPDPNPRTL-ERLEPAQRPPRRG
MUL_1313|M.ulcerans_Agy99 DRVALVTKLHHSVIDGVSGMELLGALLDTVSFPDAG-PELEKPPTGTAIG
MSMEG_4297|M.smegmatis_MC2_155 GRLALMTKVHHASVDGVTGANLLSQLCTTEPDA----PPPDP-----VDG
Mvan_3601|M.vanbaalenii_PYR-1 GRIAVMTKVHHAAVDGVTGANLMSKLCTTEPDA----PPPDP-----VAG
TH_2477|M.thermoresistible__bu GPLVVLTKVHHAAVDGVTGANLLSQVCTTEADT----PPPEP-----VQG
MAV_2240|M.avium_104 GRLAVMTKVHHAGVDGVTGANLMSQLCATEPDA----APPEP-----AAG
Mb1460|M.bovis_AF2122/97 GRIATLTKMHHAIVDGVSGAGLGEILLDITPEP----RPPQQETVG-FVG
Rv1425|M.tuberculosis_H37Rv GRIATLTKMHHAIVDGVSGAGLGEILLDITPEP----RPPQQETVG-FVG
MAB_0507|M.abscessus_ATCC_1997 GRVATLTKMHHAIVDGVSGAGLGEILLDIAPEP----RPAPDDTVGSLVG
Mflv_0349|M.gilvum_PYR-GCK GRVAVVTKMHHAVVDGVSGAGISEILLDTTPEP----RPPAVDASRSLVG
MLBr_01244|M.leprae_Br4923 GRAAMLLQISHAITDGVGSVEMFAEMYDLERDPPSRPRSPQPIPQDLSRN
. . : :: *: *** . : : . .
MMAR_1605|M.marinum_M DPVPGQVGLLLRGLLGLARQ-PLRALAATPRTLPHLDQVPTVRMLPG--V
MUL_1313|M.ulcerans_Agy99 N-ASSDAGLLARALASLPVQ-PARMVRGLPPMLRHIDQLPTMRHLPGAAV
MSMEG_4297|M.smegmatis_MC2_155 PGGGTGLEIAVNGAVRFATR-PLRLVNVVPSTLN----------------
Mvan_3601|M.vanbaalenii_PYR-1 PGGGTDLEIAVSGAVRFATR-PLKLVNVVPNTVS----------------
TH_2477|M.thermoresistible__bu AGTASDLEIALSGAVRFATR-PLKLLNVLPTTAN----------------
MAV_2240|M.avium_104 VGGGAGWRIAAGGLVRFAAR-PLRLASVVPDTAA----------------
Mb1460|M.bovis_AF2122/97 FQIPGLERRAIGALINVGIMTPFRIVRLLEQTVR----------------
Rv1425|M.tuberculosis_H37Rv FQIPGLERRAIGALINVGIMTPFRIVRLLEQTVR----------------
MAB_0507|M.abscessus_ATCC_1997 LGLPSRERILLSALVNVGVKTPYRVARLVQQTVR----------------
Mflv_0349|M.gilvum_PYR-GCK VKPPSRERQAVNGLINVWFKTPYRITRLLEQTVR----------------
MLBr_01244|M.leprae_Br4923 DLMLQGINHLPVALAGGVVGGLSGVASVAGRAILR---------------
.
MMAR_1605|M.marinum_M RALAAGSALLTGARRRDGGVLERTNLRAPRTRFNRRIGPHRRFAFASVPL
MUL_1313|M.ulcerans_Agy99 IAKVADRAERLATRNKDGRNLQHPRAAAPKVSFGGRISAQRRFAFGSLPL
MSMEG_4297|M.smegmatis_MC2_155 -------TVVDTVRRARNGRTMAAPFAAPRTAFNANITGHRNIAFAQLDL
Mvan_3601|M.vanbaalenii_PYR-1 -------TVIDTVKRARNGLTMAPPFAAPKTAFNANVTGHRNVAFAQLDL
TH_2477|M.thermoresistible__bu -------SVLDTVRRARAGRTMARPFAAPSTPFNANVTGRRNVAFTQLDL
MAV_2240|M.avium_104 -------SVLATVRRALDGAAMARPFAAPTTVFNARISNRRCIAYAELNL
Mb1460|M.bovis_AF2122/97 -------QQIAALGVAGKP---ARYFEAPKTRFNAPVSPHRRVTGTRVEL
Rv1425|M.tuberculosis_H37Rv -------QQIAALGVAGKP---ARYFEAPKTRFNAPVSPHRRVTGTRVEL
MAB_0507|M.abscessus_ATCC_1997 -------QQLAVFGMERKS---PGYFEAPHTRFNAPVSPHRRISGARLDL
Mflv_0349|M.gilvum_PYR-GCK -------QQIAVRNIDNKP---PRYFDAPKVRFNGPISPHRSVAGASVPL
MLBr_01244|M.leprae_Br4923 ----PASTVSGVVGYVRSGIRVLSQAAEPSPLLRQRSLATR-TEAIEIQL
* : * : *
MMAR_1605|M.marinum_M ADVKAVKNAM-GVTVNDVVLALSAGVLRRRLAAAGDLPTEPLLAMVPVSV
MUL_1313|M.ulcerans_Agy99 DQIKAAKFAVPGATVNDVVVAVCAGALRRRMIARGDDVATPLVAMVPVSV
MSMEG_4297|M.smegmatis_MC2_155 EDIKTVKNHF-DVKVNDVVMALVSGVLRTFLSHRDELPDNSLVAMVPVSV
Mvan_3601|M.vanbaalenii_PYR-1 EDIKTVKNHF-GVKVNDVVMALVSGVLRTFLLERGELPESSLVAMVPVSV
TH_2477|M.thermoresistible__bu EDVKRIKDHF-GVKVNDVVMALTSGALRSYLLERDQLPDSPLIAMVPVSV
MAV_2240|M.avium_104 DDVKAVKNHF-GVKVNDVVMALVSGVLRQYLAERNALPDSSLVASVPISV
Mb1460|M.bovis_AF2122/97 ARAKAVKDAF-GVKLNDVVLALVAGAARQYLQKRDELPAKPLIAQIPVST
Rv1425|M.tuberculosis_H37Rv ARAKAVKDAF-GVKLNDVVLALVAGAARQYLQKRDELPAKPLIAQIPVST
MAB_0507|M.abscessus_ATCC_1997 ARVKALKDAF-GVKVNDVVLAIVAGAVRDYLAKRDEFPDKPLIVQIPVSV
Mflv_0349|M.gilvum_PYR-GCK DRVKAVKNAF-DVKLNDVVLALVSGALRSYLKDRGELPAKSLVSQVPVST
MLBr_01244|M.leprae_Br4923 SDLHKAAKAG-DGSINDAYLAGLVGALRRYHEALG-VSISTLPMAVPVNV
: . .:**. :* *. * . .* :*:..
MMAR_1605|M.marinum_M RTPEQAGTFGNRVSTMIVPLPTDEPDARRRVARVNDVMRSAKQRHRAIPA
MUL_1313|M.ulcerans_Agy99 RTGERQ--YGNQISSMIVPIPTDQADARARLISAHQVMRAAKERHRAIPA
MSMEG_4297|M.smegmatis_MC2_155 H-DRSDRPGRNQVSGMFSPLQTHIADPAERLRAIAEANAVAKQHSSAIGA
Mvan_3601|M.vanbaalenii_PYR-1 H-DKSDRPGRNQVSGMFSRLETNIEDPATRLKAIAEANSVAKQHSSAIAA
TH_2477|M.thermoresistible__bu H-DKTDRPGRNHVSGMFSRLETTIEDPAERLRAIANANSVAKQHSSAIAA
MAV_2240|M.avium_104 H-GKSDRPGRNQVSAMFASLHTEIADPVQRLKAIAHANSLAKEHSSAIGA
Mb1460|M.bovis_AF2122/97 RSEETKADVGNQVSSMTASLATHIEDPAKRLAAIHESTLSAKEMAKALSA
Rv1425|M.tuberculosis_H37Rv RSEETKADVGNQVSSMTASLATHIEDPAKRLAAIHESTLSAKEMAKAPSA
MAB_0507|M.abscessus_ATCC_1997 RTEETDGVVGNQVSSMTVSLATDIDDPAERIQAIYEGTQGAKEMARAMSA
Mflv_0349|M.gilvum_PYR-GCK R-AQADDTVGNQVNAMTVVLATDVADPAERIKAIYSYTQGAKEMTKALTA
MLBr_01244|M.leprae_Br4923 R-TEADVVGSNRFVGVTLAAPLGTNDPAARMQKIRSQMTQRRDEPAMNII
: . *:. : *. *: ::
MMAR_1605|M.marinum_M TLMQDANQFIPPALLARASRVVGQLSVSDPLAPPVNVVVSNVPGSPTPLY
MUL_1313|M.ulcerans_Agy99 KLLTEANHTVPPALLTRAARVISMTTGSGWIRPPFNVTISNIPGSPDPLY
MSMEG_4297|M.smegmatis_MC2_155 TLLQDWTQFAAPAVFGTAMRVYAASRLS-GAKPVHNLVVSNVPGPQVPLY
Mvan_3601|M.vanbaalenii_PYR-1 TLLQDWTQFAAPAVFGAAMRVYASSRLS-GARPVHNLVISNVPGPQGPLY
TH_2477|M.thermoresistible__bu SLLQDWTQFAAPAVFGVAMRVYAASRLG-EAKPVHNLVVSNVPGPQVPLY
MAV_2240|M.avium_104 SLLQDWTQFAAPAVFGIAMRLYARTRFT-DSMPVHNLVVSNVPGPQLPLY
Mb1460|M.bovis_AF2122/97 HQIMGLTETTPPGLLQLAARAYTASGLS-HNLAPINLVVSNVPGPPFPLY
Rv1425|M.tuberculosis_H37Rv HQIMGLTETTPPGLLQLAARAYTASGLS-HNLAPINLVVSNVPGPPFPLY
MAB_0507|M.abscessus_ATCC_1997 HQIMGLTDTTPPGLLQLAARAYTASGLS-RNLAPINLVLSNVPGPSFPLY
Mflv_0349|M.gilvum_PYR-GCK HQIMGYTETTPPGLLALASRAYAASGIG-SNIAPMNVVISNVPGPTFPLY
MLBr_01244|M.leprae_Br4923 GSLAPLMTVLPASVLDFIVDSVASSDVN----------ASNIPAYPGDTY
: ...:: **:*. *
MMAR_1605|M.marinum_M LAGARLQAQFPVSIVIDGVGVNLTVLSYRDSLDIGVIADRDLVEDAWPFI
MUL_1313|M.ulcerans_Agy99 CAGARVVSQHPVNVLLDGMGLSITLLSYQDRLDFGLTADRELLPDVWHLA
MSMEG_4297|M.smegmatis_MC2_155 FLGAEVKAMYPLGPIFHGSGLNITVMSLTGKLDVGIISCPELLPDLWDMA
Mvan_3601|M.vanbaalenii_PYR-1 MLGCEVKAMYPLGPIFHGSGLNMTVMSLSGKLDVGIVSCPELLPDLWDMA
TH_2477|M.thermoresistible__bu FLGAEVKAMYPLGPIFHGSGLNITVMSLTGRLNVGMVSCPELMPDLWELA
MAV_2240|M.avium_104 LLGCEVKAMYPLGPIFHGSGLNITAMSLRGKLDVGLIGCPDLVPDLWELA
Mb1460|M.bovis_AF2122/97 MAGARLDSLVPLGPPVMDVALNITCFSYQDYLDFGLVTTPEVANDIDEMA
Rv1425|M.tuberculosis_H37Rv MAGARLDSLVPLGPPVMDVALNITCFSYQDYLDFGLVTTPEVANDIDEMA
MAB_0507|M.abscessus_ATCC_1997 MAGARLESMIPLGPPVMDVALNITCFSYLEHMDFGFVTTPEVAADIDEMA
Mflv_0349|M.gilvum_PYR-GCK LGGARVEHLMPVGPLLFDVALNVTCFSYCGAVDFGFITTPEIAADIVDLA
MLBr_01244|M.leprae_Br4923 FAGAKILRQYGIGP-RPGVAMMAVLMSRGGFCTVTVRYDRASVKSEALFA
*..: :. . .: . :* . . . :
MMAR_1605|M.marinum_M EDIKAELAELSALV-IPRPKRKTRTRTART-QPKEDPK----
MUL_1313|M.ulcerans_Agy99 DELAAELSALAAEVGAPRNPVADGAPTSTTGRPTAKPGAIPR
MSMEG_4297|M.smegmatis_MC2_155 DDFAVALDELLDATKRA-------------------------
Mvan_3601|M.vanbaalenii_PYR-1 DTFGVALQELLDATR---------------------------
TH_2477|M.thermoresistible__bu EGFERALRELLDTVG---------------------------
MAV_2240|M.avium_104 DEFAVAMEELRAAAR---------------------------
Mb1460|M.bovis_AF2122/97 DAIEPALAELERAAE---------------------------
Rv1425|M.tuberculosis_H37Rv DAIEPALAELERAAE---------------------------
MAB_0507|M.abscessus_ATCC_1997 DGLEAALVELEHAVGI--------------------------
Mflv_0349|M.gilvum_PYR-GCK DQIEPALHDLEVAAGLVSDPTRPKPRKRGRR-----------
MLBr_01244|M.leprae_Br4923 RCLLEGFDEVLALAGDPTPHAVPASFAARSSGSPAGWLSSS-
: : : .