For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSIAMPAFSPAQESLFLTLGGRALDSRLPHPFLGDTIADEILGETGYDLGKFPSLTTKLIDARTKVFDIA VRAKRLDEVAGRFIERHPDAIGLDLGAGLDSRVLRINPPSTVGWYDVDFPQIVDLRRRLLPAQVGAHNIG ADLTDPNWLDEVPADRPAVIIADGLVAFLRQHDFAALLSRLTNHFASGEIAFNLYTTQAIWATKHLASLA AISAGVVNPGFNDPRQPERWVAGLELAEEILLTRAPEVAELPLVTRLTSRLVAPSAAVSRMIGTTLVRYK F
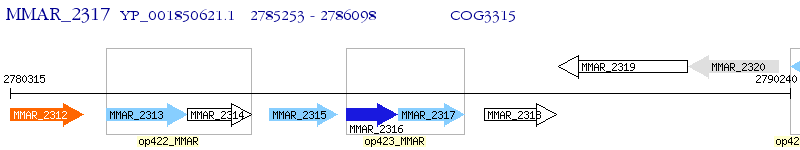
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2317 | - | - | 100% (281) | O-methyltransferase |
| M. marinum M | MMAR_2971 | - | 2e-37 | 35.66% (244) | O-methyltransferase |
| M. marinum M | MMAR_2733 | omt | 4e-18 | 27.13% (188) | O-methyltransferase Omt |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1184c | omt | 3e-16 | 27.23% (191) | O-methyltransferase |
| M. gilvum PYR-GCK | Mflv_3375 | - | 2e-45 | 36.65% (281) | O-methyltransferase domain-containing protein |
| M. tuberculosis H37Rv | Rv1153c | omt | 5e-17 | 27.75% (191) | O-methyltransferase |
| M. leprae Br4923 | MLBr_02020 | - | 2e-06 | 26.15% (218) | hypothetical protein MLBr_02020 |
| M. abscessus ATCC 19977 | MAB_1517c | - | 6e-49 | 36.14% (285) | O-methyltransferase Omt |
| M. avium 104 | MAV_4048 | - | 2e-47 | 36.49% (285) | Omt protein |
| M. smegmatis MC2 155 | MSMEG_6473 | - | 2e-37 | 33.08% (263) | tetracenomycin polyketide synthesis O-methyltransferase TcmP |
| M. thermoresistible (build 8) | TH_0783 | omt | 1e-20 | 29.10% (189) | PROBABLE O-METHYLTRANSFERASE OMT |
| M. ulcerans Agy99 | MUL_3020 | omt | 9e-17 | 26.06% (188) | O-methyltransferase Omt |
| M. vanbaalenii PYR-1 | Mvan_3107 | - | 3e-49 | 40.50% (279) | O-methyltransferase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1184c|M.bovis_AF2122/97 -------MSAHKPAKQRV-ALTGVSETALLTLNARAAEARRRDTIIDDPM
Rv1153c|M.tuberculosis_H37Rv -------MSAHKPAKQRV-ALTGVSETALLTLNARAAEARRRDAIIDDPM
TH_0783|M.thermoresistible__bu -------VHESNPDKLDASTLSGVSETALMTLYGRAEQARHPQPIIDDPM
MUL_3020|M.ulcerans_Agy99 -------MGHSD-NNVNAGMLGGVSETALLTLNGRAHQAAKPNPIIDDPM
MAB_1517c|M.abscessus_ATCC_199 -------------MGIDVSGITPEEETAFLTLKARAVNDGWRRPILRDPG
MAV_4048|M.avium_104 -------------MGIDTSAITPEEETAFLTLQARAINNGWARPILPDPG
Mflv_3375|M.gilvum_PYR-GCK ---------MCLAGGMDVSGLAPIEQTALLTEYCRALDSRSARPILGDSH
Mvan_3107|M.vanbaalenii_PYR-1 ---------MCLAGNMDVSGLTPIEQTALLTEYCRALDSKAARPILGDRL
MMAR_2317|M.marinum_M -------------MSIAMPAFSPAQESLFLTLGGRALDSRLPHPFLGDTI
MSMEG_6473|M.smegmatis_MC2_155 ------------MTDKLKVDLSGAPQTMLATFYAKALDARLPTPILGDDM
MLBr_02020|M.leprae_Br4923 MMAATPEFGSLRSDDDHWDIVSSVGYTALLVAGWRALHAVGPQPLVRDEY
. : : . :* . .:: *
Mb1184c|M.bovis_AF2122/97 AVALVESI-DFGFAKFGPTG----------QGFALRARAFDMAAQHYLDQ
Rv1153c|M.tuberculosis_H37Rv AVALVESI-DFDFAKFGPTG----------QGFALRARAFDMAAQHYLDQ
TH_0783|M.thermoresistible__bu AVRLVDRI-DFDFTKFGRRG----------QEMALRSLAFDRAALRYLSD
MUL_3020|M.ulcerans_Agy99 AIKLVESN-DFDFEKFGRRG----------QEMALRSLAVDRCEIAYLTK
MAB_1517c|M.abscessus_ATCC_199 ATAAVSKI-DYDFHALGVIT-------PVVCQSSLRAKLLDDRVRAFVTE
MAV_4048|M.avium_104 AADAVTKI-DYDFDGLGLVT-------PVVCQTSLRAKLLDDRVRAFIAQ
Mflv_3375|M.gilvum_PYR-GCK ADATVRRI-DHDFASLGATP-------SVVALVALRAKMLDECLSRFVAA
Mvan_3107|M.vanbaalenii_PYR-1 AEKTVDQI-DYDFTTLAATP-------SVVPLVALRAKMLDERLRAFTAR
MMAR_2317|M.marinum_M ADEILGET-GYDLGKFPSLTTKLIDARTKVFDIAVRAKRLDEVAGRFIER
MSMEG_6473|M.smegmatis_MC2_155 AAEIAERI-DYDWSRTAITP-------SRAPAVTTRSRHFDRWARQFLAV
MLBr_02020|M.leprae_Br4923 AKYFITASRDPYLMNLLANPGTSLNETAFPRLYGVQTRFFDDFFSSAGDT
* . :: .*
Mb1184c|M.bovis_AF2122/97 HPAATVVALAEGLQTSFWRLDVAIPGGQFRWLTVDLPPIVDLRTRLLPSS
Rv1153c|M.tuberculosis_H37Rv HPAATVVALAEGLQTSFWRLDVAIPGGQFRWLTVDLPPIVDLRTRLLPSS
TH_0783|M.thermoresistible__bu HPRATVVALAEGLQTTFWRVTDAIPDARFRWLTIDLPPIIELRERLLPAE
MUL_3020|M.ulcerans_Agy99 HPGATVVAHAEGFQTGFWRVSNALPDTQFRWVSLDLEPVMELRRRLLPDS
MAB_1517c|M.abscessus_ATCC_199 HPNAVVVDMGAGLDDGYRRIQ---PPATVDWYSVDLPGMATLRDEVMPPG
MAV_4048|M.avium_104 HADAVVVDLGAGLDDGYARVN---PPDTVDWYSVELPGLAALRDKVMPPG
Mflv_3375|M.gilvum_PYR-GCK DPSAVVVDLGAGLSSAAFRVD---PPPTVDWVNVDLPGVIGLRNAVVPAH
Mvan_3107|M.vanbaalenii_PYR-1 HPDAVVVDLGAGFSSAVFRVD---PPSTVDWYSVDLPAVIALREALLPVR
MMAR_2317|M.marinum_M HPDAIGLDLGAGLDSRVLRIN---PPSTVGWYDVDFPQIVDLRRRLLPAQ
MSMEG_6473|M.smegmatis_MC2_155 HPEAVVLHLGCGLDGRFFRLA---PGPGVEWFDVDYPEVIDLRSQLYPEH
MLBr_02020|M.leprae_Br4923 G-IRQAVIVAAGLDSRAYRLK---WPNGATVFEIDLPKVLEFKARVLAEQ
: . *:. *: :: : :: : .
Mb1184c|M.bovis_AF2122/97 PRVSVCAQS----ALDYSWMDSVDPA-----GGVFITAEGLLMYLQPEQA
Rv1153c|M.tuberculosis_H37Rv PRVSVCAQS----ALDYSWMDSVDPA-----GGVFITAEGLLMYLQPEQA
TH_0783|M.thermoresistible__bu PRIVTLAQS----ALDFSWMDRVDTD-----HGVLITAEGLLMYLQPDEA
MUL_3020|M.ulcerans_Agy99 ARVTNLAQS----ALDYTWMDKVDSC-----AAVFITAEGLLMYPQPNQA
MAB_1517c|M.abscessus_ATCC_199 PGEHTVGIS----ITDPNWLGGIPSD-----RPAMVIADGFFAFLTKEQI
MAV_4048|M.avium_104 PHEHTIGIS----VTDPEWISAIPAD-----RPAIVIADGLFPFLNKEQI
Mflv_3375|M.gilvum_PYR-GCK ERVRDVAAN----VLEPDWVTAVPPD-----RPTIVFADGLFAFLDEDAV
Mvan_3107|M.vanbaalenii_PYR-1 DRDRSVPTS----LLEPGWADSIPAD-----RPTMLFADGLVAFLTEADV
MMAR_2317|M.marinum_M VGAHNIGAD----LTDPNWLDEVPAD-----RPAVIIADGLVAFLRQHDF
MSMEG_6473|M.smegmatis_MC2_155 ARCHLVSAS----VTDPAWLDEVPTG-----RPALMLGEGLTMYLTETEG
MLBr_02020|M.leprae_Br4923 GAIPNAGRSEVAADLRADWPRALKAAGFDPQRSSAWSVEGLLPYLTNDAQ
. * : . :*: :
Mb1184c|M.bovis_AF2122/97 LGLIAQCAQTFPGG-QMLFDLPPRWFAGWSRLGLRTSLRYKVPR-----M
Rv1153c|M.tuberculosis_H37Rv LGLIAQCAQTFPGG-QMLFDLPPRWFAGWSRLGLRTSLRYKVPR-----M
TH_0783|M.thermoresistible__bu MTLIERCAARFPGG-QMLFDLPPVLLKKLAPKGLRASRRYRVPP-----M
MUL_3020|M.ulcerans_Agy99 MELISQCAKRFAGG-QMFFDLPPTIVKRVAPKGMRASRRYRVPP-----M
MAB_1517c|M.abscessus_ATCC_199 IATLRAITDHFSTG-QLAFNDYGRMVVG-VWISKLFPQKMFKKVNQLRGF
MAV_4048|M.avium_104 IAVMRAITDHFPAG-QLAFNDYGRMVIG-VWISKLFPQRMFKKVNQLRAF
Mflv_3375|M.gilvum_PYR-GCK VSLLRRITGHFPSG-VVAFNDYGRVSKANQVIGRLTSSRNANSPHRQWNF
Mvan_3107|M.vanbaalenii_PYR-1 VAILRRITGHFASG-VVAFNDYGPVSKLNRLLGRLATSSKTNSPHGQWNF
MMAR_2317|M.marinum_M AALLSRLTNHFASG-EIAFNLY---TTQAIWATKHLASLAAISAGVVN--
MSMEG_6473|M.smegmatis_MC2_155 LALLRRVVDRFGTG-ELQFDAFNTFGIRTQWTNTVVRRSGATLR------
MLBr_02020|M.leprae_Br4923 SALFTRIGELCAPGSRIAVGALGSRLDRKQLAALEATHPGVNISG-----
: * : ..
Mb1184c|M.bovis_AF2122/97 PFSMSVAQAADLVNKVPGVVAVRDLRVPPGRGLWVNMALSTVYRLPVFDP
Rv1153c|M.tuberculosis_H37Rv PFSMSVAQAADLVNKVPGVVAVRDLRVPPGRGLWVNMALSTVYRLPVFDP
TH_0783|M.thermoresistible__bu PFSLSPAQLARLADTVPGVRAVHDLPMPEGRGFFFRAVFPRFWQARPTRQ
MUL_3020|M.ulcerans_Agy99 PFSLSVNQLARLVDTVPGIRAVHDVPMPEGRGLFFRTLYPALWRFPPIKP
MAB_1517c|M.abscessus_ATCC_199 EGYNDPRTPERWNPRLRFVEEFNLVDAPEIELFPTWLRISSQLARYSTSM
MAV_4048|M.avium_104 EGYNDPHTPERWSPKLTLVEETSLASVPEVDLFPTWLRIATRMAGWSKRT
Mflv_3375|M.gilvum_PYR-GCK PGFKDARQPESWDPALRLLDESSAMHRPETAQFPTGLRIASRLSRRVPSI
Mvan_3107|M.vanbaalenii_PYR-1 PGFKDAHRPEAWNRDLTLLEEASVMQRPEGALFPAGLRLAAKLSHRVPAI
MMAR_2317|M.marinum_M PGFNDPRQPERWVAGLELAEEILLTRAPEVAELPLVTRLTSRLVAPSAAV
MSMEG_6473|M.smegmatis_MC2_155 WAIDRPDDILDTVPGTRLLAWMSVFEDPVFDELAWHYRLLAKVMRPLSPL
MLBr_02020|M.leprae_Br4923 DVDFSALTYEPKTDSAQWLAAHGWAVEPVRNTLELQTSYGMTPPDVDVQM
* :
Mb1184c|M.bovis_AF2122/97 LRPCLT-LLEFSRPARG
Rv1153c|M.tuberculosis_H37Rv LRPCLT-LLEFSRPARG
TH_0783|M.thermoresistible__bu WRGAYT-LLEFG-----
MUL_3020|M.ulcerans_Agy99 FRGAYV-LLEFD-----
MAB_1517c|M.abscessus_ATCC_199 KRSAR--ILRFEF----
MAV_4048|M.avium_104 ARSAR--ILRFSF----
Mflv_3375|M.gilvum_PYR-GCK ARKAR--VLQYRF----
Mvan_3107|M.vanbaalenii_PYR-1 ARKAR--VLQYRF----
MMAR_2317|M.marinum_M SRMIGTTLVRYKF----
MSMEG_6473|M.smegmatis_MC2_155 RYMAQY--HRYAF----
MLBr_02020|M.leprae_Br4923 DSFMHSQYITATR----