For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GHSDNDANARMLGGVSETALLTLNGRAHQAAKPNPIIDDPMAIKLVESIDFDFEKFGRRGQEMALRSLAV DRCEIAYLTKHPGATVVALAEGFQTGFWRVSNALPDTQFRWVSLDLEPVMELRRRLLPDSARVTNLAQSA LDYTWMDKVDSRAGVFITAEGLLMYLQPNQAMELISQCAKRFAGGQMFFDLPPTIVKRVAPKGMRASRRY RVPPMPFSLSVNQLARLVDSVPGIRAVHDVPMPEGRGLFFKTLYPALWRFPPIKPFRGAYVLLEFG*
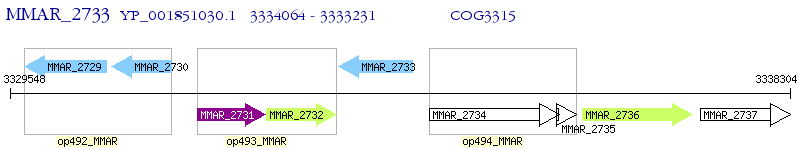
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2733 | omt | - | 100% (277) | O-methyltransferase Omt |
| M. marinum M | MMAR_4300 | omt_2 | 3e-91 | 61.13% (265) | O-methyltransferase Omt_2 |
| M. marinum M | MMAR_0262 | omt_1 | 7e-66 | 48.15% (270) | O-methyltransferase Omt_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1184c | omt | 5e-83 | 59.09% (264) | O-methyltransferase |
| M. gilvum PYR-GCK | Mflv_5259 | - | 1e-109 | 70.41% (267) | O-methyltransferase domain-containing protein |
| M. tuberculosis H37Rv | Rv1153c | omt | 8e-84 | 59.47% (264) | O-methyltransferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0668c | - | 6e-72 | 50.93% (269) | O-methyltransferase OmT |
| M. avium 104 | MAV_2991 | - | 4e-22 | 28.57% (273) | tetracenomycin polyketide synthesis O-methyltransferase TcmP |
| M. smegmatis MC2 155 | MSMEG_6661 | - | 1e-103 | 65.80% (269) | O-methyltransferase, putative |
| M. thermoresistible (build 8) | TH_0783 | omt | 1e-109 | 68.40% (269) | PROBABLE O-METHYLTRANSFERASE OMT |
| M. ulcerans Agy99 | MUL_3020 | omt | 1e-153 | 96.38% (276) | O-methyltransferase Omt |
| M. vanbaalenii PYR-1 | Mvan_0979 | - | 1e-113 | 71.17% (274) | O-methyltransferase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1184c|M.bovis_AF2122/97 ---MSAHKPAKQRVALTGVSETALLTLNARAAEARRRDTIIDDPMAVALV
Rv1153c|M.tuberculosis_H37Rv ---MSAHKPAKQRVALTGVSETALLTLNARAAEARRRDAIIDDPMAVALV
MAB_0668c|M.abscessus_ATCC_199 ---MSG----IDVRHLDGVSETALLTLHQRATEAARTDGILDDPMAISLH
MMAR_2733|M.marinum_M ---MGHSDNDANARMLGGVSETALLTLNGRAHQAAKPNPIIDDPMAIKLV
MUL_3020|M.ulcerans_Agy99 ---MGHSDNNVNAGMLGGVSETALLTLNGRAHQAAKPNPIIDDPMAIKLV
Mflv_5259|M.gilvum_PYR-GCK --------------MLAGVSETALLTLNGRAYQARHPNAILDDPMAIRVV
Mvan_0979|M.vanbaalenii_PYR-1 ----MDQDGKIDAGALAGVSETALLTLNGRAYQARHPNAIIDDPMAIRLV
MSMEG_6661|M.smegmatis_MC2_155 MDRSEEPGGKIDASALTRVSETALLTLYGRAYQGRHQQAILNDPMAIRLV
TH_0783|M.thermoresistible__bu --VHESNPDKLDASTLSGVSETALMTLYGRAEQARHPQPIIDDPMAVRLV
MAV_2991|M.avium_104 --MTATEKVDFSSVAWGSVEWTNLVTLYLRACESRSRQPILGDTAAAEAV
*. * *:** ** :. : *:.*. *
Mb1184c|M.bovis_AF2122/97 ESIDFGFAKFGP------TGQGFALRARAFDMAAQHYLDQHPAATVVALA
Rv1153c|M.tuberculosis_H37Rv ESIDFDFAKFGP------TGQGFALRARAFDMAAQHYLDQHPAATVVALA
MAB_0668c|M.abscessus_ATCC_199 ASVDYDFAHFGR------THQATALRALIFDKATREYLDYHPCATVVALA
MMAR_2733|M.marinum_M ESIDFDFEKFGR------RGQEMALRSLAVDRCEIAYLTKHPGATVVALA
MUL_3020|M.ulcerans_Agy99 ESNDFDFEKFGR------RGQEMALRSLAVDRCEIAYLTKHPGATVVAHA
Mflv_5259|M.gilvum_PYR-GCK DSIDFDFDKFGRR-----KGQEMALRSLAFDRAATAYLAKHPKATVVALA
Mvan_0979|M.vanbaalenii_PYR-1 DAIDFDFDKFGRR-----KGQEMALRSLAFDRAATRYLSRHPAATVVALA
MSMEG_6661|M.smegmatis_MC2_155 DSIEFDFEKFGR------KGQEMALRSLAFDRAALAYLSAQPRATVVALA
TH_0783|M.thermoresistible__bu DRIDFDFTKFGR------RGQEMALRSLAFDRAALRYLSDHPRATVVALA
MAV_2991|M.avium_104 DRIRYDFKRIHRMSLPASNQYLVALRAVQFDDWCAEFLARHPDAVVLHLG
:.* :: ***: .* :* :* *.*: .
Mb1184c|M.bovis_AF2122/97 EGLQTSFWRLDVAIPGGQFRWLTVDLPPIVDLRTRLLPSSPRVSVCAQSA
Rv1153c|M.tuberculosis_H37Rv EGLQTSFWRLDVAIPGGQFRWLTVDLPPIVDLRTRLLPSSPRVSVCAQSA
MAB_0668c|M.abscessus_ATCC_199 EGLQTSFWRVD----NGRLTWLSVDLEPIVRLRQRLLPRSERLSHCAQSA
MMAR_2733|M.marinum_M EGFQTGFWRVSNALPDTQFRWVSLDLEPVMELRRRLLPDSARVTNLAQSA
MUL_3020|M.ulcerans_Agy99 EGFQTGFWRVSNALPDTQFRWVSLDLEPVMELRRRLLPDSARVTNLAQSA
Mflv_5259|M.gilvum_PYR-GCK EGLQTSFWRLDAALPTAEFRWLTVDFEPIIELRRRLLPESARITTLAQSA
Mvan_0979|M.vanbaalenii_PYR-1 EGLQTSFWRLDAAIPDSRFRWVSIDLEPVIALRRRLLPDSPRITTLAQSA
MSMEG_6661|M.smegmatis_MC2_155 EGLQTTFWRLTDALPDAEFHWLTVDFQPIIDLRRRLLPADPRITCLAQSA
TH_0783|M.thermoresistible__bu EGLQTTFWRVTDAIPDARFRWLTIDLPPIIELRERLLPAEPRIVTLAQSA
MAV_2991|M.avium_104 CGLDGRAFRVSVP---PSLSWFDVDQPSVIGLRRRLYDDTENYRMIGSSV
*:: :*: : *. :* .:: ** ** . ..*.
Mb1184c|M.bovis_AF2122/97 LDYSWMDSVDPAGGVFITAEGLLMYLQPEQALGLIAQCAQTFPGGQMLFD
Rv1153c|M.tuberculosis_H37Rv LDYSWMDSVDPAGGVFITAEGLLMYLQPEQALGLIAQCAQTFPGGQMLFD
MAB_0668c|M.abscessus_ATCC_199 LDYSWMDRVDDSAGVLITAEGLFQYLERDLVFDLISVCAQRFPGGRLIFD
MMAR_2733|M.marinum_M LDYTWMDKVDSRAGVFITAEGLLMYLQPNQAMELISQCAKRFAGGQMFFD
MUL_3020|M.ulcerans_Agy99 LDYTWMDKVDSCAAVFITAEGLLMYPQPNQAMELISQCAKRFAGGQMFFD
Mflv_5259|M.gilvum_PYR-GCK LDYSWMDAVDTTHGVFITAEGLLMYLQPEESMGLITECAARFPGAQMIFD
Mvan_0979|M.vanbaalenii_PYR-1 LDYSWMDAVDTTDGVFITAEGLLMYLQPDEAMDLITACASRFPGGQMIFD
MSMEG_6661|M.smegmatis_MC2_155 LDYSWMDQVDTGDGVFITAEGLLMYLEPEESMGLIKQCANRFAGGQMVFD
TH_0783|M.thermoresistible__bu LDFSWMDRVDTDHGVLITAEGLLMYLQPDEAMTLIERCAARFPGGQMLFD
MAV_2991|M.avium_104 TELQWLDEVPAGRPTLMVAEGLLMYLREDEVRRLLQRLIDRFPGGEMHFD
: *:* * .::.****: * . : *: *.*..: **
Mb1184c|M.bovis_AF2122/97 LPPRWFAGWSRLGLRTSLRYKVPRMPFSMSVAQAADLVNKVPGVVAVRDL
Rv1153c|M.tuberculosis_H37Rv LPPRWFAGWSRLGLRTSLRYKVPRMPFSMSVAQAADLVNKVPGVVAVRDL
MAB_0668c|M.abscessus_ATCC_199 SVPKFLSTHSQRGIRLSEQYVVPPMPFSFTANQYDELR-AIPGIRSVREL
MMAR_2733|M.marinum_M LPPTIVKRVAPKGMRASRRYRVPPMPFSLSVNQLARLVDSVPGIRAVHDV
MUL_3020|M.ulcerans_Agy99 LPPTIVKRVAPKGMRASRRYRVPPMPFSLSVNQLARLVDTVPGIRAVHDV
Mflv_5259|M.gilvum_PYR-GCK LPPVMVKKFAPKGVRSSSRYRVPPMPFSLSAAELADLVHTVPGVRAVHDL
Mvan_0979|M.vanbaalenii_PYR-1 LPPTMVKKIAPKGMRSSARYRVPPMPFSLSPAQLAELVHTVPGVTAVHDL
MSMEG_6661|M.smegmatis_MC2_155 LPPVVVKKFAKKGLRSSKRYRVPPMPFSLTPNQLQDLARTVPGIRAVHDL
TH_0783|M.thermoresistible__bu LPPVLLKKLAPKGLRASRRYRVPPMPFSLSPAQLARLADTVPGVRAVHDL
MAV_2991|M.avium_104 TLSALAPLLSKVFTRG---------IITWGIRDARRIADWNPRLRFLEQS
. : * :: : : * : :.:
Mb1184c|M.bovis_AF2122/97 RVPPGRGLWVNMALSTVYRLPVFDPLRPCLTLLEFSRPARG
Rv1153c|M.tuberculosis_H37Rv RVPPGRGLWVNMALSTVYRLPVFDPLRPCLTLLEFSRPARG
MAB_0668c|M.abscessus_ATCC_199 RMPAGRGKVLRWAAPLLYALPGLDRWRAPHTVVEFG-----
MMAR_2733|M.marinum_M PMPEGRGLFFKTLYPALWRFPPIKPFRGAYVLLEFG-----
MUL_3020|M.ulcerans_Agy99 PMPEGRGLFFRTLYPALWRFPPIKPFRGAYVLLEFD-----
Mflv_5259|M.gilvum_PYR-GCK PMPQGRGFFFEKVFPAFWGLKATKNYRGAYTLLEFG-----
Mvan_0979|M.vanbaalenii_PYR-1 PMPRGRGFLFERVFPAFWQFKPTRNYRGAYTLLEFG-----
MSMEG_6661|M.smegmatis_MC2_155 PMPAGRGFLFEKVFPAFWQFKPTKQIRGAYTLLELG-----
TH_0783|M.thermoresistible__bu PMPEGRGFFFRAVFPRFWQARPTRQWRGAYTLLEFG-----
MAV_2991|M.avium_104 PALAGYQRIPSTVVRSIYRLTWLTGGRGYDVLNRFEY----
* .: * .: .: