For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALDHERLLRAAAEFLGRRPNATQDEIAAAIGVSRATLHRYFAGRAALIDALEQLAFGQMREALKSARWQE GSATEALRRLVAACESVSGYLTLLYAYSQDSDTNELKQGWLEIDSEIKELFLRGQRQGEFRPDLAAGWLT EAFYSLVSGAGWSINTGRAAPRDFAPMITELLLHGVSHSGQPNTNNHHAPERRRLGSEPTRPMINSRVSR *
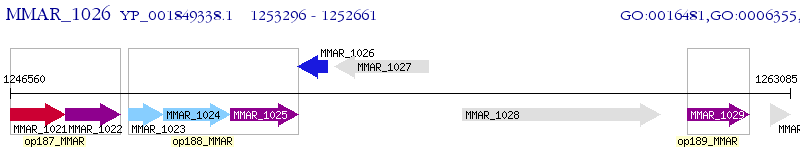
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1026 | - | - | 100% (211) | TetR family transcriptional regulator |
| M. marinum M | MMAR_2831 | - | 1e-59 | 61.67% (180) | transcriptional regulator |
| M. marinum M | MMAR_2833 | - | 6e-56 | 59.89% (182) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0240 | - | 2e-60 | 63.48% (178) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1525c | - | 6e-05 | 27.40% (146) | TetR family transcriptional regulator |
| M. avium 104 | MAV_2463 | - | 3e-07 | 28.57% (175) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6223 | - | 4e-09 | 29.79% (141) | TetR family protein transcriptional repressor LfrR |
| M. thermoresistible (build 8) | TH_2600 | - | 6e-06 | 37.50% (80) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_0778 | - | 1e-116 | 97.63% (211) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0665 | - | 6e-60 | 64.04% (178) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1026|M.marinum_M ----------------MALDHERLLRAAAEFLGRR--PNATQDEIAAAIG
MUL_0778|M.ulcerans_Agy99 ----------MMYYRLMALDHERLLRAAAEFLGRR--PNATQDEIAAAIG
Mflv_0240|M.gilvum_PYR-GCK ----------------MALSRDDLLRAAADFLGRR--PNATQDEIAAAVG
Mvan_0665|M.vanbaalenii_PYR-1 ----------------MTATRDQLLRAAADFLGRR--PNATQDEIAAAIG
MSMEG_6223|M.smegmatis_MC2_155 ------MTSPSIESGARERTRRAILDAAMLVLADH--PTAALGDIAAAAG
MAV_2463|M.avium_104 ----MTAVEDRPLRSDAARNVERILRAAREVYAELG-PDAPVEAVARRAG
TH_2600|M.thermoresistible__bu MSKPRVRNRYSSGPTRGEQRRERLLQALEDLLASRPFAAIEVGDIARAAG
MAB_1525c|M.abscessus_ATCC_199 --MSSSPSALSRVERKKREMRQEILDSAFACFAEQGYHATGVADIAGRIG
:* : . :* *
MMAR_1026|M.marinum_M VSRATLHRYFAGRAALIDALEQLAFGQMREAL--KSARWQEGSATEALRR
MUL_0778|M.ulcerans_Agy99 VSRATLHRYFAGRAALIDALEQLAFGQMREAL--KSARWQEGSATEELRR
Mflv_0240|M.gilvum_PYR-GCK VSRATLHRHFAGRPALMAALEELAVAEMRDVL--EDARLQEDSAIDALRR
Mvan_0665|M.vanbaalenii_PYR-1 VSRATLHRHFAGRVALMTALEELAITEMRTVL--DTVQLHEGTPTAALRR
MSMEG_6223|M.smegmatis_MC2_155 VGRSTVHRYYPERTDLLRALARHVHDLSNAAI--ERADPTSGPVDAALRR
MAV_2463|M.avium_104 VGERTLYRRFPAKADLVRAALDQSIAEDLTPV--IDSARHAKDPLHGLTR
TH_2600|M.thermoresistible__bu LSRSAFYFYFPSKAAAVGAMLGDVFGEISDVL--DKWTELGDSVAARRRE
MAB_1525c|M.abscessus_ATCC_199 IGHGTFYRYFKSKRDIIEHVIDDVTGQIFEAIGAENAAGLADDIEQYRQQ
:.. :.: : : : : .
MMAR_1026|M.marinum_M LVAACESVSGYL------TLLYAYSQDSDTNELKQGW--LEIDSEIKELF
MUL_0778|M.ulcerans_Agy99 LVAACESVSGYL------TLLYAYSQDSDTNELKQGW--LEIDSEIKELF
Mflv_0240|M.gilvum_PYR-GCK LVTACEPVSPYL------ALLYSQSQELDFDSTLDAW--QEIDTAITDLF
Mvan_0665|M.vanbaalenii_PYR-1 LVSACEPVSPYL------ALLYTQSQEIDMDATLEAW--QAIDDEITALF
MSMEG_6223|M.smegmatis_MC2_155 VVESQLDLGPIV------LFVYYEPSILADPELAAYF--DIGDEAIVEVL
MAV_2463|M.avium_104 LVEAAISLGARE------QNLLAAAHR-AGSLTSDIS--VSLNEALGELV
TH_2600|M.thermoresistible__bu VYDSFERVVAIWR-----QHAPLMAAMFDAASTDAEV--RAMFEGHIDSF
MAB_1525c|M.abscessus_ATCC_199 SVRIGAALAHLFRENPHLPLLLLEAGSVDSELRERVFGMLSTSDHLTEAY
: .
MMAR_1026|M.marinum_M L-RGQRQGEFRPDLAAGWLTEAFYSLVSGAGWSINTG--RAAPRDFAPMI
MUL_0778|M.ulcerans_Agy99 L-RGQRQGEFRPDLAAGWLTEAFYSLVSGAGWSINTG--RAAPRDFAPMI
Mflv_0240|M.gilvum_PYR-GCK L-RGQRDGDFRPELPAAWLTEALYGLIGGTAWSVRAG--RAAARDFNRLI
Mvan_0665|M.vanbaalenii_PYR-1 V-RGQRDGEFRPELTAAWLTEMFYSLVGGTAWSIRAG--RAAARDFDRLI
MSMEG_6223|M.smegmatis_MC2_155 N-RASTE---RPEYPPGWARRVFWALMQAGYEAAKDG--MPRHQIVDAIM
MAV_2463|M.avium_104 R-EGQRAGRIRADLVADDLPR-LVAMLYSVLSTMDAG--SEGWRRYVALV
TH_2600|M.thermoresistible__bu V-DGSAAR-IRHERAVGEAPDGPDPEALARVLVGMNV--KALERDMRAIT
MAB_1525c|M.abscessus_ATCC_199 LRHGVEKGYLRADLDTEYTARAVTGMILANGLHASRGGEVQDTERFSAAV
. * : . .
MMAR_1026|M.marinum_M TELLLHGVSHSGQPNTNNHHAPERRRLGSEPTRPMINSRVSR
MUL_0778|M.ulcerans_Agy99 TELLLHGVSHSGQPSTNNHHAPERRRLGSEPMRPMINSRATR
Mflv_0240|M.gilvum_PYR-GCK LDLLLNGATPR-------------------------------
Mvan_0665|M.vanbaalenii_PYR-1 IDLLLHGVTS--------------------------------
MSMEG_6223|M.smegmatis_MC2_155 TSLTSGIITLPRT-----------------------------
MAV_2463|M.avium_104 VDAISVDERRPLPPAAALRYASQPNSWPL-------------
TH_2600|M.thermoresistible__bu SGAPADADLPRVLAFVWGRVLYDA------------------
MAB_1525c|M.abscessus_ATCC_199 IALMYGGIGP--------------------------------