For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MALSRDDLLRAAADFLGRRPNATQDEIAAAVGVSRATLHRHFAGRPALMAALEELAVAEMRDVLEDARLQ EDSAIDALRRLVTACEPVSPYLALLYSQSQELDFDSTLDAWQEIDTAITDLFLRGQRDGDFRPELPAAWL TEALYGLIGGTAWSVRAGRAAARDFNRLILDLLLNGATPR
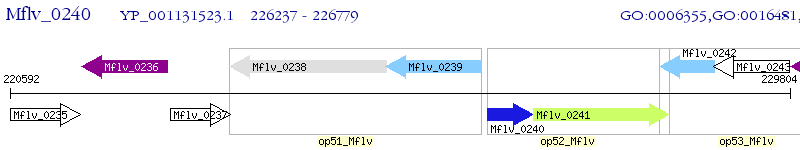
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0240 | - | - | 100% (180) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4113 | - | 4e-06 | 34.95% (103) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2228 | - | 5e-05 | 28.47% (137) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3185c | - | 1e-07 | 29.05% (148) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3160c | - | 2e-07 | 29.05% (148) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1026 | - | 2e-60 | 63.48% (178) | TetR family transcriptional regulator |
| M. avium 104 | MAV_2463 | - | 3e-06 | 26.79% (168) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6223 | - | 6e-10 | 30.86% (162) | TetR family protein transcriptional repressor LfrR |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0778 | - | 6e-60 | 62.92% (178) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0665 | - | 2e-75 | 76.40% (178) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0240|M.gilvum_PYR-GCK ------------MALSRDDLLRAAADFLGRR--PNATQDEIAAAVGVSRA
Mvan_0665|M.vanbaalenii_PYR-1 ------------MTATRDQLLRAAADFLGRR--PNATQDEIAAAIGVSRA
MMAR_1026|M.marinum_M ------------MALDHERLLRAAAEFLGRR--PNATQDEIAAAIGVSRA
MUL_0778|M.ulcerans_Agy99 ------MMYYRLMALDHERLLRAAAEFLGRR--PNATQDEIAAAIGVSRA
Mb3185c|M.bovis_AF2122/97 -----MPRQAGRWSPTALRILGAAAELIALRGYSSTSTRDIAAAVGVEQP
Rv3160c|M.tuberculosis_H37Rv -----MPRQAGRWSPTALRILGAAAELIALRGYSSTSTRDIAAAVGVEQP
MSMEG_6223|M.smegmatis_MC2_155 --MTSPSIESGARERTRRAILDAAMLVLADH--PTAALGDIAAAAGVGRS
MAV_2463|M.avium_104 MTAVEDRPLRSDAARNVERILRAAREVYAELG-PDAPVEAVARRAGVGER
:* ** . . . :. :* ** .
Mflv_0240|M.gilvum_PYR-GCK TLHRHFAGRPALMAALEELAVAEMRDVLEDARLQEDSAIDALRRLVTACE
Mvan_0665|M.vanbaalenii_PYR-1 TLHRHFAGRVALMTALEELAITEMRTVLDTVQLHEGTPTAALRRLVSACE
MMAR_1026|M.marinum_M TLHRYFAGRAALIDALEQLAFGQMREALKSARWQEGSATEALRRLVAACE
MUL_0778|M.ulcerans_Agy99 TLHRYFAGRAALIDALEQLAFGQMREALKSARWQEGSATEELRRLVAACE
Mb3185c|M.bovis_AF2122/97 AIYKHFSAKRDILAALVRLAVEWPLELFGHITAMPVPAVVKLHRWLTESL
Rv3160c|M.tuberculosis_H37Rv AIYKHFSAKRDILAALVRLAVEWPLELFGHITAMPVPAVVKLHRWLTESL
MSMEG_6223|M.smegmatis_MC2_155 TVHRYYPERTDLLRALARHVHDLSNAAIERADPTSGPVDAALRRVVESQL
MAV_2463|M.avium_104 TLYRRFPAKADLVRAALDQSIAEDLTPVIDSARHAKDPLHGLTRLVEAAI
:::: :. : :: * . * * :
Mflv_0240|M.gilvum_PYR-GCK P---VSPYLALLYSQSQELDFD---STLDAWQEIDTAITDLFLRGQRDGD
Mvan_0665|M.vanbaalenii_PYR-1 P---VSPYLALLYTQSQEIDMD---ATLEAWQAIDDEITALFVRGQRDGE
MMAR_1026|M.marinum_M S---VSGYLTLLYAYSQDSDTN---ELKQGWLEIDSEIKELFLRGQRQGE
MUL_0778|M.ulcerans_Agy99 S---VSGYLTLLYAYSQDSDTN---ELKQGWLEIDSEIKELFLRGQRQGE
Mb3185c|M.bovis_AF2122/97 DHLHASPYVLVSILITPDLHQESFVAERELVAEMERALVGLIETGQGEGD
Rv3160c|M.tuberculosis_H37Rv DHLHASPYVLVSILITPDLHQESFVAERELVAEMERALVGLIETGQGEGD
MSMEG_6223|M.smegmatis_MC2_155 D---LGPIVLFVYYEPSILADP---ELAAYFDIGDEAIVEVLNRASTE--
MAV_2463|M.avium_104 S---LGAREQNLLAAAHRAGSL----TSDISVSLNEALGELVREGQRAGR
. . : : :. ..
Mflv_0240|M.gilvum_PYR-GCK FR--PELPAAWLTEALYGLIGGTAWSVRAGRAAARDFNRLILDLLLNGAT
Mvan_0665|M.vanbaalenii_PYR-1 FR--PELTAAWLTEMFYSLVGGTAWSIRAGRAAARDFDRLIIDLLLHGVT
MMAR_1026|M.marinum_M FR--PDLAAGWLTEAFYSLVSGAGWSINTGRAAPRDFAPMITELLLHGVS
MUL_0778|M.ulcerans_Agy99 FR--PDLAAGWLTEAFYSLVSGAGWSINTGRAAPRDFAPMITELLLHGVS
Mb3185c|M.bovis_AF2122/97 VRAMHPLSAARLVQALFDALALPEFAVSPDEIVEFAMTALLSDPDRLAEI
Rv3160c|M.tuberculosis_H37Rv VRAMHPLSAARLVQALFDALALPEFAVSPDEIVEFAMTALLSDPDRLAEI
MSMEG_6223|M.smegmatis_MC2_155 -R--PEYPPGWARRVFWALMQAGYEAAKDGMPRHQIVDAIMTSLTSGIIT
MAV_2463|M.avium_104 IR--ADLVADDLPR-LVAMLYSVLSTMDAGSEGWRRYVALVVDAISVDER
* . . : : : . :: .
Mflv_0240|M.gilvum_PYR-GCK PR-------------------------------
Mvan_0665|M.vanbaalenii_PYR-1 S--------------------------------
MMAR_1026|M.marinum_M HSGQPNTNNHHAPERRRLGSEPTRPMINSRVSR
MUL_0778|M.ulcerans_Agy99 HSGQPSTNNHHAPERRRLGSEPMRPMINSRATR
Mb3185c|M.bovis_AF2122/97 RAAADALEIQTAPPDRGL---------------
Rv3160c|M.tuberculosis_H37Rv RAAADALEIQTAPPDRGL---------------
MSMEG_6223|M.smegmatis_MC2_155 LPRT-----------------------------
MAV_2463|M.avium_104 RPLPPAAALRYASQPNSWPL-------------