For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ETERLLRAAADHLSTRPNATLDEIGAAAGVSHSTLYRHFDGRTALLEALDHAAIEQMRDALKTSHWQEYS PIDALRILVAACEPVAGYLTLRYVQGQSFETRKSVAEWREINSEIEEIFLRGQRAGEFRTDVTADWLTEA FFSLVSGAGWSVQHGRVARREFTHMITALFLDGVKAPTQGNLPPPLVGFVLLGLSVVRPNLPNSKS*
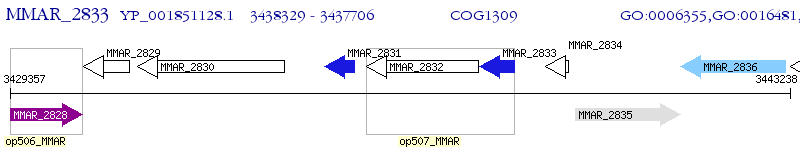
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2833 | - | - | 100% (207) | TetR family transcriptional regulator |
| M. marinum M | MMAR_2830 | - | e-107 | 98.95% (191) | hypothetical protein MMAR_2830 |
| M. marinum M | MMAR_1026 | - | 6e-56 | 59.89% (182) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0240 | - | 4e-47 | 52.30% (174) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2061c | - | 3e-05 | 43.64% (55) | TetR family transcriptional regulator |
| M. avium 104 | MAV_2463 | - | 9e-09 | 27.57% (185) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_6223 | - | 3e-07 | 26.90% (171) | TetR family protein transcriptional repressor LfrR |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2922 | - | 1e-88 | 98.17% (164) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0665 | - | 1e-46 | 53.49% (172) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2833|M.marinum_M ------------METER--LLRAAADHLSTR--PNATLDEIGAAAGVSHS
MUL_2922|M.ulcerans_Agy99 ------------METER--LLRAAADFLSTR--PNATLDEIGAAAGVSHS
Mflv_0240|M.gilvum_PYR-GCK ------------MALSRDDLLRAAADFLGRR--PNATQDEIAAAVGVSRA
Mvan_0665|M.vanbaalenii_PYR-1 ------------MTATRDQLLRAAADFLGRR--PNATQDEIAAAIGVSRA
MSMEG_6223|M.smegmatis_MC2_155 --MTSPSIESGARERTRRAILDAAMLVLADH--PTAALGDIAAAAGVGRS
MAV_2463|M.avium_104 MTAVEDRPLRSDAARNVERILRAAREVYAELG-PDAPVEAVARRAGVGER
MAB_2061c|M.abscessus_ATCC_199 -----------MAGLRRKLLLEAAADLFARQGFHAVGIDDIGAAAGVSGP
:* ** . . :. **.
MMAR_2833|M.marinum_M TLYRHFDGRTALLEALDHAAIEQMRDALKTSHWQEYSPIDALRILVAACE
MUL_2922|M.ulcerans_Agy99 TLYRHFDGRTALLEALDHAAIEQMRDALKTSHWQEYSPTDALRILVAACE
Mflv_0240|M.gilvum_PYR-GCK TLHRHFAGRPALMAALEELAVAEMRDVLEDARLQEDSAIDALRRLVTACE
Mvan_0665|M.vanbaalenii_PYR-1 TLHRHFAGRVALMTALEELAITEMRTVLDTVQLHEGTPTAALRRLVSACE
MSMEG_6223|M.smegmatis_MC2_155 TVHRYYPERTDLLRALARHVHDLSNAAIERADPTSGPVDAALRRVVESQL
MAV_2463|M.avium_104 TLYRRFPAKADLVRAALDQSIAEDLTPVIDSARHAKDPLHGLTRLVEAAI
MAB_2061c|M.abscessus_ATCC_199 AVYRHFQNKDAILRELCDAAMTELLAGARRATT-AGTPTEVLHALVDLHA
:::* : : :: * :*
MMAR_2833|M.marinum_M PVAGY---LTLRYVQGQSFETRKSVAEWREINSEIEEIFLRG--QRAGEF
MUL_2922|M.ulcerans_Agy99 PVAGY---LTLRYVQGQSFETRKSVAEWREINSEIEEIFLRG--QRAGEF
Mflv_0240|M.gilvum_PYR-GCK PVSPY---LALLYSQSQELDFDSTLDAWQEIDTAITDLFLRG--QRDGDF
Mvan_0665|M.vanbaalenii_PYR-1 PVSPY---LALLYTQSQEIDMDATLEAWQAIDDEITALFVRG--QRDGEF
MSMEG_6223|M.smegmatis_MC2_155 DLGPI---VLFVYYEPSILADPELAAYFDIGDEAIVEVLNRA--STE---
MAV_2463|M.avium_104 SLGAR---EQNLLAAAHRAGS-LTSDISVSLNEALGELVREG--QRAGRI
MAB_2061c|M.abscessus_ATCC_199 AFGARRRGLLAVYAREHRSLSPLATRALRRRQHSYESFWVEALVRARGDL
.. : . ..
MMAR_2833|M.marinum_M RTDVTADWLTEAFFSLVSGAGWSVQHGRVARREFTHMITALFLDGVKAPT
MUL_2922|M.ulcerans_Agy99 RTDVTADWLTEAFFSLVSGAGWSVQLGRVARREFT---------------
Mflv_0240|M.gilvum_PYR-GCK RPELPAAWLTEALYGLIGGTAWSVRAGRAAARDFNR--------------
Mvan_0665|M.vanbaalenii_PYR-1 RPELTAAWLTEMFYSLVGGTAWSIRAGRAAARDFDR--------------
MSMEG_6223|M.smegmatis_MC2_155 RPEYPPGWARRVFWALMQAGYEAAKDGMPRHQIVDA--------------
MAV_2463|M.avium_104 RADLVADDLPR-LVAMLYSVLSTMDAGSEGWRRYVALVVDAISVDERRP-
MAB_2061c|M.abscessus_ATCC_199 DRERAQGLVAAVLSLLNASAYMPASLDDATIEAMLAAAAVA---------
: : : . . . .
MMAR_2833|M.marinum_M QGNLPPPLVGFVLLGLSVVRPNLPNSKS-----
MUL_2922|M.ulcerans_Agy99 ------------------ARRANPAWRD-----
Mflv_0240|M.gilvum_PYR-GCK ----------------LILDLLLNGATPR----
Mvan_0665|M.vanbaalenii_PYR-1 ----------------LIIDLLLHGVTS-----
MSMEG_6223|M.smegmatis_MC2_155 -----------------IMTSLTSGIITLPRT-
MAV_2463|M.avium_104 ---------------LPPAAALRYASQPNSWPL
MAB_2061c|M.abscessus_ATCC_199 ---------------ALFSRAVTQQVDGAPHRI