For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDHRRPTERDAIMRAAYRLISEGAAGPSASSSTSIENILRVARVNRRVFYRHFASKDDLIIAMQEWAGDL ILADLQKAVAAADTPAAAVAAWIEDYLSIGWQEARFRDALAFMSAEVIGAPGIAAALEVTYDRHRAVLTE ALAAGLADGSLPNAHPELDSFAIHAVTVRHLEARIRGRLDSDFREVRDQIVTLMLTGLSAPAVYRTATR*
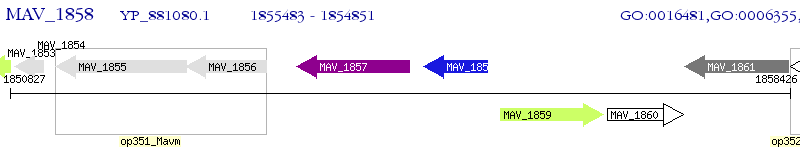
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1858 | - | - | 100% (210) | transcriptional regulator, TetR family protein |
| M. avium 104 | MAV_2439 | - | 6e-10 | 30.12% (166) | transcriptional regulator, TetR family protein, putative |
| M. avium 104 | MAV_0309 | - | 8e-08 | 31.87% (91) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2097 | - | 2e-18 | 36.81% (163) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2913 | - | 3e-06 | 25.37% (205) | TetR family regulatory protein |
| M. marinum M | MMAR_1569 | - | 2e-09 | 27.92% (197) | putative regulatory protein |
| M. smegmatis MC2 155 | MSMEG_1355 | - | 1e-07 | 28.87% (194) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3734 | - | 3e-18 | 29.90% (194) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_3464 | - | 2e-10 | 28.93% (197) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_4608 | - | 8e-18 | 35.15% (165) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1569|M.marinum_M MSDIAATMKCRLRVSRKRLVGTYGSMTHPNNPGTRRQADLSSRLTRRRHK
MUL_3464|M.ulcerans_Agy99 MSDIAATMKCRLRVSRKRLVGTYGSMTHPNNPGTRRQADLSSRLTRRRHK
Mflv_2097|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4608|M.vanbaalenii_PYR-1 -----------------------------------------------MQP
TH_3734|M.thermoresistible__bu ---------------------------------------------VAETR
MAV_1858|M.avium_104 ------------------------------------------------MS
MAB_2913|M.abscessus_ATCC_1997 ------------------------------MYPYRFTVITTGRLLMSTGS
MSMEG_1355|M.smegmatis_MC2_155 --------------------------------------MTTPTRWAGVPL
MMAR_1569|M.marinum_M LAPDPGIRRAILVAASMSVREHGV-----RGLSVATVLDRAQLSTRAFYR
MUL_3464|M.ulcerans_Agy99 LAPDPGIRRAILVAASMSVREHGV-----RGLSVATVLDRAQLSTRAFYR
Mflv_2097|M.gilvum_PYR-GCK -----------MEAAYRCLADPHA-----GPVSMAAILGTAGVSSRAFYR
Mvan_4608|M.vanbaalenii_PYR-1 GPDHDGDRRRIIEAAYRCLAQPHA-----GPIPMTSILRSAGLSSRAFYR
TH_3734|M.thermoresistible__bu TVDRSSERQHIIDAAYRCLDRNSG-----SSASITEILDEAGLGTRAFYR
MAV_1858|M.avium_104 DHRRPTERDAIMRAAYRLISEGAAGPSASSSTSIENILRVARVNRRVFYR
MAB_2913|M.abscessus_ATCC_1997 TRRREHTRARLMQAALDVFADRGF-----HGASIENICEKAGFTRGAFYS
MSMEG_1355|M.smegmatis_MC2_155 TDRRAERRALLVDAAFRLFGDGGE-----TALSVRSVCRECGLNTRYFYE
: ** . .: : . . **
MMAR_1569|M.marinum_M HFESKDQLVAAVFLEVARVEAVRLRTLMAKAT-NPIEGVAAWIDGRLDLA
MUL_3464|M.ulcerans_Agy99 HFESKDQLVAAMFLEVARVEAVRLRTLMAKAT-NPIEGVAAWIDGRLDLA
Mflv_2097|M.gilvum_PYR-GCK HFESKDDLFLALLEQECSAVVGQVDRVADAAQGSPADQLAAWIEAMFDIV
Mvan_4608|M.vanbaalenii_PYR-1 HFLSKDDLFLALLRQECEAVIAQVDRIADAAVGSPADQLADWIAAMFDII
TH_3734|M.thermoresistible__bu NFSTKHDLLLEMFRRDREAVQAQLREAVARAP-TPVEALRSWMTHMFELV
MAV_1858|M.avium_104 HFASKDDLIIAMQEWAGDLILADLQKAVAAAD-TPAAAVAAWIEDYLSIG
MAB_2913|M.abscessus_ATCC_1997 NFAGKDELFFTLFDASSEQLLTQLRAALDECR-KSPDPLSTFIRTIDDKG
MSMEG_1355|M.smegmatis_MC2_155 SFADTDDLIGAVYDEVSALLAADVEAAMEGAGNSLRARTRAGIAAVLGFS
* ..:*. : : . . :
MMAR_1569|M.marinum_M FDDAVKADLRHLSMDAQSHLLNTPELVEPAYAETLEPLIEQLHLGVAQEV
MUL_3464|M.ulcerans_Agy99 FDDAVKADLRHLSMGAQSHLLNAPELVEPAYAETLEPLIEQLHLGVAQEV
Mflv_2097|M.gilvum_PYR-GCK VDPRQRLQLTVIDSEEVRTAKGYRQTRDRLHAARERGLAEILRRGLRDGS
Mvan_4608|M.vanbaalenii_PYR-1 VDPQQRLQLTVIDSDEVRAAKGYREMRERLHAERERSLAEILRRGRRDGS
TH_3734|M.thermoresistible__bu SNPRKRRRVATLYSEEMRRTPGYVRELELVTAADQASIAAILHRGKAEGI
MAV_1858|M.avium_104 WQEARFRDALAFMSAEVIGAPGIAAALEVTYDRHRAVLTEALAAGLADGS
MAB_2913|M.abscessus_ATCC_1997 PTQRRWYLISMEFTLYAIREPSAAVVLAEHDARLRREAAGIINELMSIAH
MSMEG_1355|M.smegmatis_MC2_155 SADPRRGRVLFTDARANPVLAQRRAVTQDLLREAVLSEGGRLNPGS----
:
MMAR_1569|M.marinum_M FHDVDPHSEARSIQ-GVVWANTEQQWASGDCRRDEVRESTVR-FCLRGLG
MUL_3464|M.ulcerans_Agy99 FHDVDPHSEARSIQ-GVVWANTEQQWASGDCRRDEVRESTVR-FCLRGLG
Mflv_2097|M.gilvum_PYR-GCK FPLTEPEADAVAIN-AVVSRVLNTQTTDEPEARKKALVAVLD-FALRAVG
Mvan_4608|M.vanbaalenii_PYR-1 FPLAEPEIDAVAVN-ALVSRLLVRQTTDDPQARKQAEVAVLD-FALRAVG
TH_3734|M.thermoresistible__bu FTACTPEADARSIHAAFEAALLDRFHQKSTTDAADEVEHLLA-FFLRGLG
MAV_1858|M.avium_104 LPNAHPELDSFAIHAVTVRHLEARIRGRLDSDFREVRDQIVT-LMLTGLS
MAB_2913|M.abscessus_ATCC_1997 RELIIDTELIARLATALREGVAAQFYVEPELAETRMLERLAFPAMLRAFS
MSMEG_1355|M.smegmatis_MC2_155 -DPVAAQVGAAMYTGAMAELAQQWLAGNLGDDLDAVVDHALR-LVLGR--
*
MMAR_1569|M.marinum_M VAPDTIAQITAPD
MUL_3464|M.ulcerans_Agy99 VAPDTIAQITAPD
Mflv_2097|M.gilvum_PYR-GCK AAAR---------
Mvan_4608|M.vanbaalenii_PYR-1 AVPGGQDTRR---
TH_3734|M.thermoresistible__bu VAT----------
MAV_1858|M.avium_104 APAVYRTATR---
MAB_2913|M.abscessus_ATCC_1997 QPVGAD-------
MSMEG_1355|M.smegmatis_MC2_155 -------------