For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SALAGGVFASGTVEDDGAELRQLVDDIGRRSFDAKLGRRRVPDEFDADLWRNLEDTGLARLTSTPDLDAG PHELAIVLYGLARHAGAVPLAETDALAGWLGREAGIELPNGPLTVAIADSDLAGGRLTGTAMDVPWSGAA TAVLLAARGRDGSRVCVLDGGETSADDGYNLAGEPRGRLTFDVAADELTPVDDAVADELSRRGGWCRCIQ IVGALDAAAALTVAHTRERVQFGRPLSAFQAVQQSLAGMAGEIERARAAVELAVAAAAEHGFGSPHTDYA VTVAKVAVGRAVIPVTTTAHQLHGAIGVTSEHPLWLFTLRAQSWADDYGTTAHHARRLGRMALAAGDPWD LVIGNLTRPKTVDK*
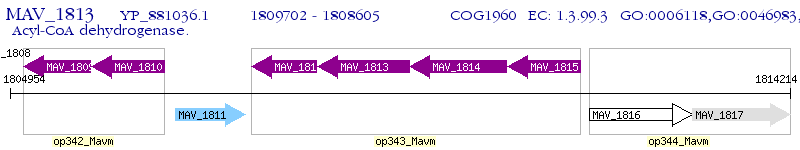
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1813 | - | - | 100% (365) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_2553 | - | e-152 | 73.28% (363) | acyl-CoA dehydrogenase, C- |
| M. avium 104 | MAV_1631 | - | 1e-25 | 30.41% (342) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3535 | fadE27 | 1e-17 | 26.90% (368) | acyl-CoA dehydrogenase FADE27 |
| M. gilvum PYR-GCK | Mflv_5167 | - | 8e-51 | 37.77% (323) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3505 | fadE27 | 7e-18 | 27.62% (362) | acyl-CoA dehydrogenase FADE27 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 5e-12 | 40.00% (105) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0594c | - | 3e-19 | 29.14% (302) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_3512 | - | 2e-45 | 36.31% (314) | acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2878 | - | 6e-47 | 36.66% (311) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4629 | - | 1e-131 | 67.43% (350) | PUTATIVE putative acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_2756 | - | 3e-45 | 35.99% (314) | acyl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2875 | - | 1e-51 | 39.75% (317) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3535|M.bovis_AF2122/97 ----------MDFTTTEAAQDLGGLVDTIVDAVCTPEHQRELDKLEQRFD
Rv3505|M.tuberculosis_H37Rv ----------MDFTTTEAAQDLGGLVDTIVDAVCTPEHQRELDKLEQRFD
MAV_1813|M.avium_104 -MSALAGGVFASGTVEDDGAELRQLVDDIGRRSFDAKLG--RRRVPDEFD
TH_4629|M.thermoresistible__bu --------VFATDTA-DDYTDLRQLVDDIGRRSFDARIG--TRRVPDQFD
MMAR_3512|M.marinum_M ----------MSATVHTVDPALVDMMEAVFAQYRTSH----PPTEASRRD
MUL_2756|M.ulcerans_Agy99 ----------MSATVHTVDPALVNMMEAVFAQYRTSH----PPTEASRRD
MSMEG_2878|M.smegmatis_MC2_155 ---------------MTVDAELVAMMNAVFADHRTAH----GPTAG----
Mflv_5167|M.gilvum_PYR-GCK -----------MTGPTSVDPDLVEMLDAVFADHREQLG---AQRPAATWD
Mvan_2875|M.vanbaalenii_PYR-1 ------MTDVHADPGMAVDADLLEMMDAVLADYRQGH----PPAGVVRFD
MAB_0594c|M.abscessus_ATCC_199 ----------MKFDLDTQQRDFAASIDAALSVADVPAAARAWAAGNHGPG
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAA--DVDQRARFP
: :
Mb3535|M.bovis_AF2122/97 RELWRKLIDAGILSSAAPESLGGDGFGVLEQVAVLVALGHQLAAVPYLES
Rv3505|M.tuberculosis_H37Rv RELWRKLIDAGILSSAAPESLGGDGFGVLEQVAVLVALGHQLAAVPYLES
MAV_1813|M.avium_104 ADLWRNLEDTGLARLTSTPD---LDAGPHELAIVLYGLARHAGAVPLAET
TH_4629|M.thermoresistible__bu DELWGNLSDTGLTRLTSDPD---LGAGPNELAVVLSGIARYAGAVPLAET
MMAR_3512|M.marinum_M PALWQRLEQLGLARLTGAEDSGGSGASWYEAAELLAAAVRNAVRIPLAEH
MUL_2756|M.ulcerans_Agy99 PALWQRLEQLGLARLTGAEDSGGSGASWYEAAELLAAAVRNAVRIPLAEH
MSMEG_2878|M.smegmatis_MC2_155 PELWDQLRALGLARLTGAEESGGSGAGWHEAAELMAAAVRSGVRVGLPEH
Mflv_5167|M.gilvum_PYR-GCK AGLWDRLGQLGLTRLTGSEESGGSGAGWREGAELLRAAVQNGVQVPLAEH
Mvan_2875|M.vanbaalenii_PYR-1 AELWESLHQLGLVRLTGAPGSGGSGAGWAESAALISSATRHAVRVPLAEH
MAB_0594c|M.abscessus_ATCC_199 LAVWSTLAELGVTALAVSEKYDGIGAHPVDLVVALEGLGKWAVPGPVVES
MLBr_00737|M.leprae_Br4923 EEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPA
* *. . : :
Mb3535|M.bovis_AF2122/97 V-VLAAGALARFGSPELQQGWGVSAVSGDRILTVALDGEMGEGPVQAAGT
Rv3505|M.tuberculosis_H37Rv V-VLAAGALARFGSPELQQGWGVSAVSGDRILTVALDGEMGEGPVQAAGT
MAV_1813|M.avium_104 D-ALAGWLGREAGIE-----------LPNGPLTVAIADSDLAG-------
TH_4629|M.thermoresistible__bu D-ALAGWLGREAGLD-----------LPDGPLTVAIAEHSGDG-------
MMAR_3512|M.marinum_M D-LLACWLLDAHDMG-----------ADAAVRTVCLLDA-----------
MUL_2756|M.ulcerans_Agy99 D-LLACWLLDSHDMG-----------ADAAVRTVCLLDA-----------
MSMEG_2878|M.smegmatis_MC2_155 D-LLACWLLEVAGIA-----------VDDNTRTVALLDE-----------
Mflv_5167|M.gilvum_PYR-GCK D-LLACWLLESAGLP-----------VDDARRTVCVLD------------
Mvan_2875|M.vanbaalenii_PYR-1 D-LLACWLLEAAGLP-----------VDDAVRTVCVLDE-----------
MAB_0594c|M.abscessus_ATCC_199 V-AVAPVLLSSHEDGAEPS-------------------------------
MLBr_00737|M.leprae_Br4923 VNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRT
:.
Mb3535|M.bovis_AF2122/97 -----GHGYRLTGTRTQVGYGPVADAFLVPAETDSGAAV------FLVAA
Rv3505|M.tuberculosis_H37Rv -----GHGYRLTGTRTQVGYGPVADAFLVPAETDSGAAV------FLVAA
MAV_1813|M.avium_104 --------GRLTGTAMDVPWSGAATAVLLAARG-RDGSR------VCVLD
TH_4629|M.thermoresistible__bu -----------PVKAVDVPWARAAAAVLLAVRT-PDGAR------IGITS
MMAR_3512|M.marinum_M -----------DGVATEVPWASHADRIVVIRQS-AGEYH------LADVD
MUL_2756|M.ulcerans_Agy99 -----------DGVATEVPWASRADRIVVIRQS-AGEYR------LADVD
MSMEG_2878|M.smegmatis_MC2_155 -----------SGTATAVPWASTAQRIVLVWRTQTGEHR------VADVD
Mflv_5167|M.gilvum_PYR-GCK -----------DGVAHGVPWASEADRIVVVWRT-NGTAQ------VADVA
Mvan_2875|M.vanbaalenii_PYR-1 -----------AGTATAVPWAAYAGRVVAIWSD-GDSHR------VADID
MAB_0594c|M.abscessus_ATCC_199 -----------TRLASGELIATVAAPPIAPRALDADIAG------LVLIA
MLBr_00737|M.leprae_Br4923 RAKADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIVHKD
. : . .
Mb3535|M.bovis_AF2122/97 GDPGVAVTALATTGLG--SVGHLELNGAKVDAARRVGGT----DVVVWLG
Rv3505|M.tuberculosis_H37Rv GDPGVAVTALATTGLG--SVGHLELNGAKVDAARRVGGT----DVAVWLG
MAV_1813|M.avium_104 GGETSADDGYNLAGE---PRGRLTFDVAADEL--TPVDD----AVADELS
TH_4629|M.thermoresistible__bu --NPQVVDGHNAAGE---PRGRVTVDLGAVTL--ASVDD----AVLTELT
MMAR_3512|M.marinum_M AQTLTITPGANMIGE---PRNTVRADHLDLRG--VPTTG----AVVSTLK
MUL_2756|M.ulcerans_Agy99 AQTLTITPGANMIGE---PRNTVRADHLDLRG--VPTAG----AVVSTLK
MSMEG_2878|M.smegmatis_MC2_155 AAAVRITAGANMIGE---PRDTVSAD-VPADG--ATVSE----ATVTTLH
Mflv_5167|M.gilvum_PYR-GCK AGDLAVTSGANLAGE---PRDTVAADPAALTG--TTVPN----ALIDQLL
Mvan_2875|M.vanbaalenii_PYR-1 RGSLHIAPGTNMIGE---PRDTVTADISALAPHSTAIDA----DLVEQLR
MAB_0594c|M.abscessus_ATCC_199 DDIGIREAQAGAQHES--IDATRRVFDVQPKEAGISLDT----SRAIDFG
MLBr_00737|M.leprae_Br4923 DEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTALATL
Mb3535|M.bovis_AF2122/97 TLSTLSRTAFQLGVLERGLQMTAEYARTREQFDRPIGSFQAVGQRLADGY
Rv3505|M.tuberculosis_H37Rv TLSTLSRTAFQLGVLERGLQMTAEYARTREQFDRPIGSFQAVGQRLADGY
MAV_1813|M.avium_104 RRGGWCRCIQIVGALDAAAALTVAHTRERVQFGRPLSAFQAVQQSLAGMA
TH_4629|M.thermoresistible__bu LRGAWCRAVQIVGALDTAAAMTVRHTAEREQFGRSLSKFQAVQHALAGMA
MMAR_3512|M.marinum_M FKSALVRAIQVCAALDQILHLSVEHVSSRVQFGRPLAKFQAVQNLIADIA
MUL_2756|M.ulcerans_Agy99 FKSALVRAIQVCAALDQILHLSVEHVSSRVQFGRPLAKFHAVQNLIADIA
MSMEG_2878|M.smegmatis_MC2_155 LKAALVRAVQVCAAMDTILELAVEHSSSRIQFGRPLAKFQAIQHMISDIA
Mflv_5167|M.gilvum_PYR-GCK LRGALVRGLQVCAALDRIVALSIAHTTDRTQFGRPLAKFQSVQNLVSDSA
Mvan_2875|M.vanbaalenii_PYR-1 LKCALVRAIQVCSALDTILVLAVEHTTSRVQFGRSLSKFQAVQHMIADMA
MAB_0594c|M.abscessus_ATCC_199 ---VLAISAQLIGAGQALLDRAVAYAKQRSQFGRPIGSYQAVKHKLADVH
MLBr_00737|M.leprae_Br4923 DHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLADMA
. : : : * **...:. :::: ::.
Mb3535|M.bovis_AF2122/97 IDVKGLRLTLTQAAWRVAEDSLASRECPQPADIDVATAGFWAAEAGHRVA
Rv3505|M.tuberculosis_H37Rv IDVKGLRLTLTQAAWRVAEDSLASRECPQPADIDVATAGFWAAEAGHRVA
MAV_1813|M.avium_104 GEIERARAAVELAVAAAAEHGFGS----PHTDYAVTVAKVAVGRAVIPVT
TH_4629|M.thermoresistible__bu GEIERARAAASLAVAAAADYGFAA----PRTEFAITAARVTAGRAVTAVT
MMAR_3512|M.marinum_M AETALARAATEAALTKAVTSQWSA----QNLDFLVAVARSCVGHAVSVVV
MUL_2756|M.ulcerans_Agy99 AETALARAATEAALTKAVTSQWSA----QNLDFLVAVARSCVGHVVSVVV
MSMEG_2878|M.smegmatis_MC2_155 AEATLARTATEAALTAAVDTTWSA----PHLEFLVAVARSCCGHAASVVV
Mflv_5167|M.gilvum_PYR-GCK AEAALARAATEAALTEAVRTGWSG----TNLEFLVAVARSCTGHATSVVV
Mvan_2875|M.vanbaalenii_PYR-1 GEAALASASTEAALAAAIASGWAA----DNLGFLVAVARSCTGHAASVVV
MAB_0594c|M.abscessus_ATCC_199 IALELARPLVYGAALSLADQSPTA-------ARDASAAKVAAAKAAYLAA
MLBr_00737|M.leprae_Br4923 MKVEAARLIVYAAAARAERGEPDLG-------FISAASKCFASDIAMEVT
* :.: . ..
Mb3535|M.bovis_AF2122/97 HTIVHVHGGVGVDTDHPVHRYFLAAKQTEFALGGATGQLRRIGRELAETP
Rv3505|M.tuberculosis_H37Rv HTIVHVHGGVGVDTDHPVHRYFLAAKQTEFALGGATGQLRRIGRELAETP
MAV_1813|M.avium_104 TTAHQLHGAIGVTSEHPLWLFTLRAQSWADDYGTTAHHARRLGRMALAAG
TH_4629|M.thermoresistible__bu TSAHQLHGAIGVTAEHPLWSVTLRAQAWADEFGTTAGYARRLGRLALESA
MMAR_3512|M.marinum_M RNAHQVHGAIGTTEEHRLHEFTRAALAWRSEFGSVHYWDEQVTKAALHAG
MUL_2756|M.ulcerans_Agy99 RNAHQVHGAIGTTEEHRLHEFTRAALAWRSEFGSVHYWDEQVTKAALHAG
MSMEG_2878|M.smegmatis_MC2_155 RNAHQVFGAIGTTAEHRLHEYTRAALAWRGEFGSVRSWDERVTTAALQAG
Mflv_5167|M.gilvum_PYR-GCK RNAHQVHGAIGTTREHRLHEFTQPALAWRSEFGSVHYWDELLTTAALAAG
Mvan_2875|M.vanbaalenii_PYR-1 RNAHQALGAIGTTIEHRLHEYTRAALTWRSEYGSVQHWDEVVTRNALDAG
MAB_0594c|M.abscessus_ATCC_199 RSALQTHGAIGFTAEYDLSLWILKVRALTSAWGTSEWHRARVLEAL----
MLBr_00737|M.leprae_Br4923 TDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR--
: *. * :. : . * :
Mb3535|M.bovis_AF2122/97 A-------------------
Rv3505|M.tuberculosis_H37Rv A-------------------
MAV_1813|M.avium_104 DP--WDLVIGNLTRPKTVDK
TH_4629|M.thermoresistible__bu NP--WDLVIGNV--------
MMAR_3512|M.marinum_M GSGLWGLITG----------
MUL_2756|M.ulcerans_Agy99 GSGLWGLITG----------
MSMEG_2878|M.smegmatis_MC2_155 GAGLWELITS----------
Mflv_5167|M.gilvum_PYR-GCK RDGLWPLITA----------
Mvan_2875|M.vanbaalenii_PYR-1 TRGLWGLITG----------
MAB_0594c|M.abscessus_ATCC_199 --------------------
MLBr_00737|M.leprae_Br4923 --------------------