For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQAIVATSYGGADVLEFRDITTPGPGPGQVLINVKAAGVNPIDWKLYSGAFGTDPDKLPMRLGLEISGTI AAVGAGVDGLVPGDDVIAAGQIGGYATRVIAAADQVFKKPASLSFNEAAGFLLTGQTAVHLLEATNVTEG DTVLIHGAAGGVGLLATQLAKARGATVVATASAARHDQLRGYGALPVEYGPGLQERVSAIGPVDAALDLV GTDEAADVSLALVADKGRIATIAGFGRAASDGFKALGGGPGADPGTEIRLAARPELIRLAGNGELKVTVD RTYPLSDARQAHEYGQTGHARGKIILLP*
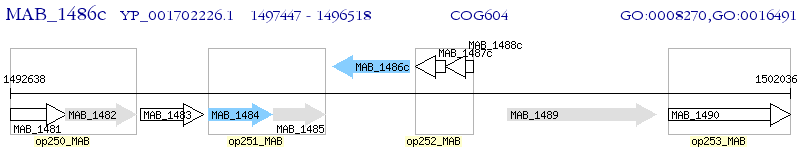
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1486c | - | - | 100% (309) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0750 | - | 4e-51 | 37.90% (314) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1874 | - | 2e-44 | 37.15% (323) | putative oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1489c | qor | 1e-32 | 34.88% (344) | quinone reductase |
| M. gilvum PYR-GCK | Mflv_2124 | - | 7e-36 | 34.69% (320) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv1454c | qor | 1e-32 | 34.88% (344) | quinone reductase |
| M. leprae Br4923 | MLBr_00118 | - | 1e-23 | 28.31% (325) | putative oxidireductase |
| M. marinum M | MMAR_2950 | - | 2e-32 | 31.72% (331) | oxidoreductase |
| M. avium 104 | MAV_5196 | - | 8e-42 | 34.41% (311) | oxidoreductase, zinc-binding dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_6747 | - | 6e-56 | 39.42% (312) | oxidoreductase |
| M. thermoresistible (build 8) | TH_3440 | - | 3e-37 | 34.90% (341) | PUTATIVE Alcohol dehydrogenase, zinc-binding domain protein |
| M. ulcerans Agy99 | MUL_2209 | - | 2e-33 | 32.02% (331) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_2724 | - | 6e-37 | 36.80% (337) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1489c|M.bovis_AF2122/97 -----------------------------------------------MHA
Rv1454c|M.tuberculosis_H37Rv -----------------------------------------------MHA
TH_3440|M.thermoresistible__bu VQTAAAQRRDEIGVIGVALRAVRTTGQNDPRMEHIRAGELHTGMIGAMKA
Mvan_2724|M.vanbaalenii_PYR-1 -----------------------------------------------MYA
MAV_5196|M.avium_104 ------------------------------------------MTAAAARA
MSMEG_6747|M.smegmatis_MC2_155 ----------------------------------------------MPRT
MAB_1486c|M.abscessus_ATCC_199 ----------------------------------------------MTQA
MMAR_2950|M.marinum_M -----------------------------------------------MKA
MUL_2209|M.ulcerans_Agy99 -----------------------------------------------MKA
Mflv_2124|M.gilvum_PYR-GCK -------------------------------------------MSDRMKA
MLBr_00118|M.leprae_Br4923 -----------------------------------------------MHA
:
Mb1489c|M.bovis_AF2122/97 IEVTETGGPGVLRHVDQPQPQPGHGELLIKAEAIGVNFIDTYFRSG----
Rv1454c|M.tuberculosis_H37Rv IEVTETGGPGVLRHVDQPQPQPGHGELLIKAEAIGVNFIDTYFRSG----
TH_3440|M.thermoresistible__bu IEVRETGGPEVLTYVDVPEPTPGPGQVLIKAEAIGVNFIDTYFRSG----
Mvan_2724|M.vanbaalenii_PYR-1 IEVAETGGPEVLSFVERPQPTPGPGQVLIKAEAIGVNFIDTYFRSG----
MAV_5196|M.avium_104 VRFDRYGGREVLYVADIEMPAPGPGEVVVEVRAAGINPGEAAIRVG----
MSMEG_6747|M.smegmatis_MC2_155 VQFAEYGDTDVLTVVDTPAPEPGPGQVRLKVRAAGLNPIDWKIVAG----
MAB_1486c|M.abscessus_ATCC_199 IVATSYGGADVLEFRDITTPGPGPGQVLINVKAAGVNPIDWKLYSG----
MMAR_2950|M.marinum_M MAQRTWNPAEPLELIELPTPEPKAGEVRVQVQAIGVNPVDWKMRSSGPLR
MUL_2209|M.ulcerans_Agy99 MAQRTWNPAEPLEFIELPTPEPQAGEVRVQVQAIGVNPVDWKMRSSGPLR
Mflv_2124|M.gilvum_PYR-GCK VVLARFGGADAFELRDVAVPHVGPRQVRVRVHATAVNPLDYQIRRG----
MLBr_00118|M.leprae_Br4923 IVAESAN---QMVWREVPDVTAGPGEVLIEVAASGVNRADVLQVAG----
: . : : :: :.. * .:* : .
Mb1489c|M.bovis_AF2122/97 ----QYPRELPFVIGSEVCGTVEAVGPGVTAADTAISVGDRVVSAS----
Rv1454c|M.tuberculosis_H37Rv ----QYPRELPFVIGSEVCGTVEAVGPGVTAADTAISVGDRVVSAS----
TH_3440|M.thermoresistible__bu ----LYPREVPFIVGTEVCGTVEAVGDGV----AALRVGDRVLTDK----
Mvan_2724|M.vanbaalenii_PYR-1 ----LYPREVPFVVGTEVCGVIEAVGEGV----AALKVGDRVVTDK----
MAV_5196|M.avium_104 ALHERFPATFPSGEGSDLAGVVTAVGPGVT----EFAVGDEVLGFS----
MSMEG_6747|M.smegmatis_MC2_155 FMRELMPVDLPAGVGSDVAGIVDAVGPGVT----EWAVGNEVLGRS----
MAB_1486c|M.abscessus_ATCC_199 AFGTD-PDKLPMRLGLEISGTIAAVGAGVD----GLVPGDDVIAAG----
MMAR_2950|M.marinum_M LAARLIGPKPPVVVGVDFAGVVEAVGPGVT----RAAPGDRVVGGTNFSR
MUL_2209|M.ulcerans_Agy99 LAARLIGPKPPVVVGVDFAGVVEAVGPGVT----RAAPGDRVVGGTNFSR
Mflv_2124|M.gilvum_PYR-GCK --DYTDLVPLPAIIGHDISGVIEEVGSHVT----EFRAGDAVYYTP-KIF
MLBr_00118|M.leprae_Br4923 --KYPHPPGASAIIGMEVAGVVAAVSSGVT----EWSVGQEVCALL----
. * :..* : *. * *: *
Mb1489c|M.bovis_AF2122/97 -ANGAYAEFCTAPASLTAKVPDDVTSEVAASALLKGLTAHYLLKSVYP--
Rv1454c|M.tuberculosis_H37Rv -ANGAYAEFCTAPASLTAKVPDDVTSEVAASALLKGLTAHYLLKSVYP--
TH_3440|M.thermoresistible__bu -ANGAYAEYTVAPADYVAYVPDGVAPDTVASALLKGMTAHYLIKSVYE--
Mvan_2724|M.vanbaalenii_PYR-1 -SLGAYAEYCLAPADFVAYVPDGVAPEVAASALLKGMTAHYLIKSTYP--
MAV_5196|M.avium_104 LRRSSHATHTAVPVGQLIRKPPELSWEVAGSLYVVGVTAYAAVRAVA---
MSMEG_6747|M.smegmatis_MC2_155 ATG-AYAEYALAEAAELIGKPGEITWEVAGSLAGAGGTAHAVLDRLD---
MAB_1486c|M.abscessus_ATCC_199 QIG-GYATRVIAAADQVFKKPASLSFNEAAGFLLTGQTAVHLLEATN---
MMAR_2950|M.marinum_M GQRGSYADTVLVREDQLCRVPDTVDIAVAAALPVGGVTAWMAVVELGRIR
MUL_2209|M.ulcerans_Agy99 GQRGSYADTVLVREDQLCRVPDTVDIAVAAALPVGGVTAWMAVVELGRIR
Mflv_2124|M.gilvum_PYR-GCK GGPGSYAEQHVADVDLVGRKPDNLDHLEAASLTLVGGTVWESLVTRAQ--
MLBr_00118|M.leprae_Br4923 -AGGGYAEYVAVPASQVMPIPKGVNVVDSAGIPEVACTVWSNLVMAAH--
.:* . * : .. . *. :
Mb1489c|M.bovis_AF2122/97 --VKRGDTVLVHAGAGGVGLILTQWATHLGVRVITTVSTAEKAKLSKDAG
Rv1454c|M.tuberculosis_H37Rv --VKRGDTVLVHAGAGGVGLILTQWATHLGVRVITTVSTAEKAKLSKDAG
TH_3440|M.thermoresistible__bu --VQRGDTILVHAGAGGVGLILTQWATHLGARVITTVSTPQKAELSRKAG
Mvan_2724|M.vanbaalenii_PYR-1 --VQQGDVVLLHAGAGGVGLILTQWATSLAASVITTVSTPDKAELSRRAG
MAV_5196|M.avium_104 --PQPGETVAVSAAAGGVGSLVTQLLVLRKARVLGIAGP-GNADWLRAHG
MSMEG_6747|M.smegmatis_MC2_155 --LKSGETLLVHAAAGGVGTFAVQLAGARGVSVIGTAGE-GNHDYLRSIG
MAB_1486c|M.abscessus_ATCC_199 --VTEGDTVLIHGAAGGVGLLATQLAKARGATVVATASA-ARHDQLRGYG
MMAR_2950|M.marinum_M QVPPADRRVLVLGASGGVGQLAVQIAKLQEAFVVGVCST-KNVEMVKDLG
MUL_2209|M.ulcerans_Agy99 QVPSADRRVLVLGASGGVGQLAVQIAKLQEAFVVGVCST-ENVEMVKDLG
Mflv_2124|M.gilvum_PYR-GCK --LTVGETILIHGGAGGVGTIAIQVAKAMGARVITTAKA-GDHAFVNSLG
MLBr_00118|M.leprae_Br4923 --LSAGQLLLLHGGTSGIGSHGIQVARALGAKVAVTAGSEEKLEFCRALG
. : : ..:.*:* * . * . *
Mb1489c|M.bovis_AF2122/97 ADVVLDYPEDAWQFAGRVRELTGGTGVQAVYDGVGATTFDASLASLAVRG
Rv1454c|M.tuberculosis_H37Rv ADVVLDYPEDAWQFAGRVRELTGGTGVQAVYDGVGATTFDASLASLAVRG
TH_3440|M.thermoresistible__bu AAEVLDYPTDPEEFGARIRDLTGGEGVPVVYDGVGRSTFDASLASLAVRG
Mvan_2724|M.vanbaalenii_PYR-1 AVEVLDYPDDPKAFGDRIRELTDGDGVAAVFDGVGASTFEASLASLAVRG
MAV_5196|M.avium_104 VVPIAYGDG-----LTDRLREAAPDGIDAFIDLFGPQYVQ-LAVDLGVPP
MSMEG_6747|M.smegmatis_MC2_155 ATPVTYGDG-----LLERVRAAAPQGVDAVLDASGRGEIP-LSIELTGDP
MAB_1486c|M.abscessus_ATCC_199 ALPVEYGPG-----LQERVSAIGP--VDAALDLVGTDEAADVSLALVADK
MMAR_2950|M.marinum_M ADLVLDYNQ-----GDALAQAKPHAPFHVVIDCVGSYSGAGCRALLSSRG
MUL_2209|M.ulcerans_Agy99 ADLVLDYNQ-----GDALAQAKPHAPFHVVIDCVGSYSGAGCRALLSSRG
Mflv_2124|M.gilvum_PYR-GCK ADVAIDHTS--TDYVDAVAEATRGRGVDVVFDTIGGDTLTRSPLSLADSG
MLBr_00118|M.leprae_Br4923 AQITINYRDE----DFVARLQQQNGGADVILDIMGAAYLDRNIDALAIDG
. . * * *
Mb1489c|M.bovis_AF2122/97 TLALFGAASGPVPPVDP----QRLNAAGSVYLTRPSLFHFTRTGEEFSWR
Rv1454c|M.tuberculosis_H37Rv TLALFGAASGPVPPVDP----QRLNAAGSVYLTRPSLFHFTRTGEEFSWR
TH_3440|M.thermoresistible__bu TLALFGAASGPVPPFDP----QRLNAAGSVYLTRPSLVHFTRTPDEFGWR
Mvan_2724|M.vanbaalenii_PYR-1 TLALFGAASGPVPPVDP----QRLNAAGSVYLTRPNLAHYTRTPDEFSWR
MAV_5196|M.avium_104 ERIDTIISFE--------------KAAEVGAKTEGSAEASTP------EV
MSMEG_6747|M.smegmatis_MC2_155 QRVLTLVAFD--------------QAGTGIQVHAGGAGDSLS------KA
MAB_1486c|M.abscessus_ATCC_199 GRIATIAGFG--------------RAASDGFKALGGGPGADPGTEIRLAA
MMAR_2950|M.marinum_M RHIMVAGDEP--------------SSALQVVVPPFKSKAILGRPN--GAR
MUL_2209|M.ulcerans_Agy99 RHIMVAGDEP--------------SSAVQVVVPPFKSKAILGRPN--GAR
Mflv_2124|M.gilvum_PYR-GCK RVVSIVDIAR---------------PQNLIEAWGRNAAYHFVFTRQNRGK
MLBr_00118|M.leprae_Br4923 QLIVIGLQGGVKGELNLGKVLSKRARIIGSTLRARPVNGPNSKSEIVAAV
Mb1489c|M.bovis_AF2122/97 AAELFDAIGSEAITVAVGGRYPLADALRAHQDLEARK-----------TV
Rv1454c|M.tuberculosis_H37Rv AAELFDAIGSEAITVAVGGRYPLADALRAHQDLEARK-----------TV
TH_3440|M.thermoresistible__bu AGEFLSAVADGTLTITVGERYPLAEAERAHRDLEGRK-----------TV
Mvan_2724|M.vanbaalenii_PYR-1 AGELLTAIADGTLDVTVSHRYALRDAEQAHRDLQGRK-----------TV
MAV_5196|M.avium_104 LAEMADLIVSGALEFDIAATYPLDRVADAFAELEQRH-----------TH
MSMEG_6747|M.smegmatis_MC2_155 LSDVVALITDGRLTVPISATYPLDEAAAALDASRTGH-----------AT
MAB_1486c|M.abscessus_ATCC_199 RPELIRLAGNGELKVTVDRTYPLSDARQAHEYGQTGH-----------AR
MMAR_2950|M.marinum_M LEPLVDAVAAGKLRVSIAQKLPLTSAEEAHRLSQTSR-----------MT
MUL_2209|M.ulcerans_Agy99 LEPLVDAVAAGKLRVSIAQKLPLTSAEEAHRLSQTSR-----------MT
Mflv_2124|M.gilvum_PYR-GCK LDALTMLVERGLVKPVLGATLPLARMGEAHELLENRRSYALRGKVAIDVA
MLBr_00118|M.leprae_Br4923 TASIWPMIADGSVRPIIGARMAVQQAADAHQLLLSGK-----------VS
. : : .: * :
Mb1489c|M.bovis_AF2122/97 GSVVLLP-------------
Rv1454c|M.tuberculosis_H37Rv GSVVLLP-------------
TH_3440|M.thermoresistible__bu GSVVLLP-------------
Mvan_2724|M.vanbaalenii_PYR-1 GSVVLLP-------------
MAV_5196|M.avium_104 GKIVLLPTDSQ---------
MSMEG_6747|M.smegmatis_MC2_155 GKIVVVP-------------
MAB_1486c|M.abscessus_ATCC_199 GKIILLP-------------
MMAR_2950|M.marinum_M GKLILEP-------------
MUL_2209|M.ulcerans_Agy99 GKLILEP-------------
Mflv_2124|M.gilvum_PYR-GCK GETVALPPRIS---------
MLBr_00118|M.leprae_Br4923 GKIVLTVAGSGTETLLSRHH
*. :