For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAVLQSYITGKWTTPEGDADPLVNAATGDEVARYAVGSVDLAAVVAYGREVGGPALRALTFHQRAAALKA LAKQLMAAKNDFYPLSAATGATARDSAVDIDGGFGTLFSYASKGVRELPNDTIYLDGASESLGRAGTFVG QHIYTSRPGVAVQINAFNFPVWGMLEKLAPAFLAGVPSIVKPAHQTAYLTELVFRAIIDSGLLPEGSVQL LCVRSHELLDHLDGQDTVLFTGSAATAAKLRSHRNVVAGGVRLNAEADSLNCSILGPDATPGTPEFDLYI KQLVAEMTTKAGQKCTAIRRALVPEALVDAAVTATSERLSKVVVGNPELDSVRMGALASIDQRDEVLRSL KLLQDSATLVFGDPASFTVEGADAAQGAFLPPLLLRAEDPTAAAVHEVEAFGPVSTVIGYRDLDDAVELA ARGQGSLVGSLVTHDPEVARKVTIGIAAHHGRILVLDRDDAKESTGHGSPLPTLVHGGPGRAGGGEELGG IRGVLHFMQRTAIQASPDMLTAITGRWTTGSARTATEVHPFRKHLEELQIGDTIVGGPRVVSLEDIDHFA EFTGDTFYAHTDPDAAARNPLFGGIVAHGYLVVSLAAGLFVEPNPGPVLANFGVDGLRFLTPVKPGDALT VTLTAKQLTPRLSADYGEVRWDAVVTNQNDDPVATYDVLTLVAKKES*
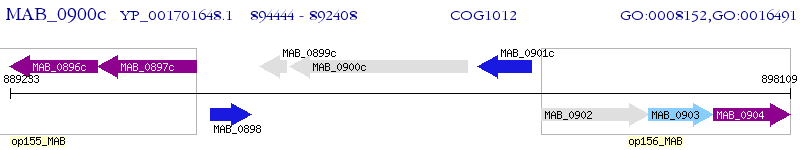
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0900c | - | - | 100% (678) | bifunctional aldehyde dehydrogenase/enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 4e-29 | 29.62% (503) | gamma-aminobutyraldehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4484 | - | 5e-27 | 28.20% (461) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0791 | aldA | 1e-19 | 30.10% (299) | aldehyde dehydrogenase NAD dependant AldA |
| M. gilvum PYR-GCK | Mflv_4860 | - | 1e-28 | 28.66% (499) | gamma-aminobutyraldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0223c | - | 5e-26 | 27.98% (511) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 6e-17 | 25.78% (481) | succinic semialdehyde dehydrogenase |
| M. marinum M | MMAR_4246 | aldA_1 | 3e-26 | 28.18% (511) | NAD-dependent aldehyde dehydrogenase, AldA_1 |
| M. avium 104 | MAV_1340 | - | 1e-30 | 30.16% (514) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1665 | - | 3e-29 | 27.71% (498) | gamma-aminobutyraldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_3902 | - | 2e-27 | 28.57% (497) | PUTATIVE putative 5-carboxymethyl-2-hydroxymuconate |
| M. ulcerans Agy99 | MUL_0939 | aldA_1 | 6e-27 | 28.18% (511) | NAD-dependent aldehyde dehydrogenase, AldA_1 |
| M. vanbaalenii PYR-1 | Mvan_1572 | - | 4e-25 | 27.38% (493) | gamma-aminobutyraldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4860|M.gilvum_PYR-GCK -----------------MTVASSWIDGAPVKTGGAQHTVINPATGESVAE
Mvan_1572|M.vanbaalenii_PYR-1 ------------------MLAGSWIDGAPVNTRGPVHQVINPATGAPVAE
MSMEG_1665|M.smegmatis_MC2_155 ---------------MAVKVASSWINGASVETSGETHQIIDPATGKSVSE
MLBr_02573|M.leprae_Br4923 ---------------------------MSIAT-------INPATGETVKT
MMAR_4246|M.marinum_M ----MAERALPE--SLVDTYQLYIDGRWVEPENGR-YDDVSPSTGMVIAT
MUL_0939|M.ulcerans_Agy99 ----MAERALPE--SLVDTYQLYIDGRWVEPENGR-YDDVSPSTGMVIAT
MAV_1340|M.avium_104 ----MAD---PP--ALVQTYQLYIDGQWVEPQDGR-YDDLSPCTEAVIAT
Mb0791|M.bovis_AF2122/97 ----MALWG-------DGISALLIDGKLSDGRAGT-FPTVNPATEEVLGV
TH_3902|M.thermoresistible__bu ---LRANRFQPKDNTVTWKYDLYIDGKWDTGTSGRALTVVNPATEETIGT
Rv0223c|M.tuberculosis_H37Rv MSD-----------SATEYDKLFIGGKWTKPSTSDVIEVRCPATGEYVGK
MAB_0900c|M.abscessus_ATCC_199 ---------------MTAVLQSYITGKWTTPE-GDADPLVNAATGDEVAR
..* :
Mflv_4860|M.gilvum_PYR-GCK FALAQPGDVDRAVVSARAALPG--WKGATPAERSAVLAKLAKLADDATAD
Mvan_1572|M.vanbaalenii_PYR-1 YALAQPADVDAAVASARAALGG--WATATPAERSAVLAKLAKLADEATAD
MSMEG_1665|M.smegmatis_MC2_155 YNLATAADVDTAVAAARAAFPA--WAGATPAERSAVLAKLAKLADEHAEQ
MLBr_02573|M.leprae_Br4923 FTPTTQDEVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQ
MMAR_4246|M.marinum_M APDASVSQVGDAVDAARRAFDAGPWPAMSHQERGHCLAQLGAALQDYADA
MUL_0939|M.ulcerans_Agy99 APDASVSQVGDAVDAARRAFDAGPWPAMSHQERGHCLAQLGAALQDYADA
MAV_1340|M.avium_104 APDASVAQVDDAIAAARRAFDSGPWGRISAEQRARCLNQLGEALTKHADD
Mb0791|M.bovis_AF2122/97 AADADAEDMGRAIEAARRAFDSTDWSRN-TELRVRCVRQLRDAMQQHVEE
TH_3902|M.thermoresistible__bu VPDASLDDIRRAIAAARTAFDEGPWPCMTPLERAAVIRRIAEVLQRRRSE
Rv0223c|M.tuberculosis_H37Rv VPMAAAADVDAAVAAARAAFDNGPWPSTPPHERAAVIAAAVKMLAERKDL
MAB_0900c|M.abscessus_ATCC_199 YAVGSVD--LAAVVAYGREVGGPALRALTFHQRAAALKALAKQLMAAKND
*: . *
Mflv_4860|M.gilvum_PYR-GCK LVAEEVSQTGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKASAEYS---
Mvan_1572|M.vanbaalenii_PYR-1 LVAEEVSQTGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKASGEYS---
MSMEG_1665|M.smegmatis_MC2_155 FVAEEVSQTGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKATAEYS---
MLBr_02573|M.leprae_Br4923 SAALMTLEMGKTLQSAKDEAMK-CAKGFRYYAEKAETLLADEPAEAAAV-
MMAR_4246|M.marinum_M YFALSQVEWGCTANERR-LQIDA-SASAARRAGHLAGTLGDQPIQSASG-
MUL_0939|M.ulcerans_Agy99 YFTLSQVEWGCTANERR-LQIDA-SASAARRAGHLASTLGDQPIQSASG-
MAV_1340|M.avium_104 FFALSQTEWGCIGNERI-MQIDG-AGFMSMHAAQLATQLVDQEVAGISAG
Mb0791|M.bovis_AF2122/97 LRELTISEVGAPRMLTASAQLEGPVGDLSFAADTAESYPWKQDLGEASPL
TH_3902|M.thermoresistible__bu LVDLVRAETGSPQWIVEMLQVGVPLRVCADVADRVLPQFRFVEPMLPSFD
Rv0223c|M.tuberculosis_H37Rv FTKLLAAETGQPPTIIETMHWMGSMGAMNYFAGAADKVTWTETRTGSYG-
MAB_0900c|M.abscessus_ATCC_199 FYPLSAATGATAR--DSAVDIDGGFGTLFSYASKGVRELPNDTIYLDGAS
. *
Mflv_4860|M.gilvum_PYR-GCK -------GDHTSSIRREAVGVVATITPWNYPLQMAVWKVIPALAAGCSVV
Mvan_1572|M.vanbaalenii_PYR-1 -------ADHTSSIRREAVGVVATITPWNYPLQMAVWKVLPALAAGCSVV
MSMEG_1665|M.smegmatis_MC2_155 -------GDHTSSIRREAVGVVATITPWNYPLQMAVWKVIPALAAGCAVV
MLBr_02573|M.leprae_Br4923 -------GASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGL
MMAR_4246|M.marinum_M ----------AAVLCHEPLGVVSILTPWNFPHSLNVMKLSRALAGGNTVV
MUL_0939|M.ulcerans_Agy99 ----------AAVLCHEPLGVVSILTPWNFPHSLNVMKLSRALAGGNTVV
MAV_1340|M.avium_104 ----------TTLLRHVPLGVVAVLTPWNFPHCLNVMKLNHALAAGNTVV
Mb0791|M.bovis_AF2122/97 G------IATRRTLAREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVV
TH_3902|M.thermoresistible__bu L------RPGQGIISKEPIGVAALITPFNFPLFLNVMKLAPALAAGCTVV
Rv0223c|M.tuberculosis_H37Rv ----------QSIVSREPVGVVGAIVAWNVPLFLAVNKIAPALLAGCTIV
MAB_0900c|M.abscessus_ATCC_199 ESLGRAGTFVGQHIYTSRPGVAVQINAFNFPVWGMLEKLAPAFLAGVPSI
**. : .:* * : *: .* :
Mflv_4860|M.gilvum_PYR-GCK IKPAEITPLTTLTLARLASEAG-LPDGVLNVVTGSGADVGAALAGHADVD
Mvan_1572|M.vanbaalenii_PYR-1 IKPAEITPLTTLTLARLAAEAG-LPAGVFNVVTGSGADVGTALAGHADVD
MSMEG_1665|M.smegmatis_MC2_155 IKPAEITPLTTLTLARLASEAG-LPDGVFNVVTGSGAEVGTALAGHRDVD
MLBr_02573|M.leprae_Br4923 LKHASNVPQCALYLADVIARGG-FPEDCFQTLLISANGVEAILR-DPRVA
MMAR_4246|M.marinum_M LKPSPLTPLAGLALARIIHEHTDIPPGVVNVVTPGGIEASKVLAVDPRID
MUL_0939|M.ulcerans_Agy99 LKPSPLTPLAGLALARIIHEHTDIPPGVVNVVTPGGIEASKVLTVDPRID
MAV_1340|M.avium_104 LKPSPLTPLAGLALARLIDEHTDIPPGVVNVVTPSGVEAAKLLTTDPRID
Mb0791|M.bovis_AF2122/97 LKPAPDTPWCAAALGEIIVEHTDFPPGVVNIVTSSSHALGALLAKDPRVD
TH_3902|M.thermoresistible__bu LKPSPLTPLEALVLGEVADEVG-LPPGVLNIVT-GGAEESAELTTNPMVD
Rv0223c|M.tuberculosis_H37Rv LKPAAETPLTANALAEVFAEVG-LPEGVLSVVP-GGIETGQALTSNPDID
MAB_0900c|M.abscessus_ATCC_199 VKPAHQTAYLTELVFRAIIDSGLLPEGSVQLLCVRSHELLDHLD---GQD
:* : .. : :* . .. : . *
Mflv_4860|M.gilvum_PYR-GCK MVTFTGSTPVGRRVMLAAAAHGHRTQLELGGKAPFVVFDDADLDAAI---
Mvan_1572|M.vanbaalenii_PYR-1 MVTFTGSTPVGRKVMLAAAAHGHRTQLELGGKAPFVVFDDADLDAAI---
MSMEG_1665|M.smegmatis_MC2_155 VVTFTGSTAVGRKVMASAAVHGHRTQLELGGKAPFVVFSDADLDAAV---
MLBr_02573|M.leprae_Br4923 AATLTGSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAV---
MMAR_4246|M.marinum_M MVSFTGSSAVGREVMAAAAGTMKRVLLECGGKSASIVLDDAECSDELLA-
MUL_0939|M.ulcerans_Agy99 MVSFTGSSAVGCEVMAAAAGTMKRVLLECGGKSASIVLDDAECSDELLA-
MAV_1340|M.avium_104 MVSFTGSSAVGRQVMSAAGDTIKRILLELGGKSASIVLDDANVTDEMLE-
Mb0791|M.bovis_AF2122/97 MISFTGSTATGRAVMADAAATIKKVFLELGGKSAFVVLDDADLAAAS-A-
TH_3902|M.thermoresistible__bu LVSFTGSDTVGRQVMNQASATLKKVVLELGGKSANVIFADADLDAAA---
Rv0223c|M.tuberculosis_H37Rv MFTFTGSSAVGREVGRRAAEMLKPCTLELGGKSAAIILEDVDLAAAI---
MAB_0900c|M.abscessus_ATCC_199 TVLFTGSAATAAKLRSHRNVVAGGVRLNAEADSLNCSILGPDATPGTPEF
:*** ... : *: .. : . :
Mflv_4860|M.gilvum_PYR-GCK ----QGAVAGALINTGQDCTAATRAIVAEGLYDDFVAGVAEVMSKVVIGD
Mvan_1572|M.vanbaalenii_PYR-1 ----QGAVAGALINTGQDCTAATRAIVARDLYDDFVAGVAEVMSNIVVGD
MSMEG_1665|M.smegmatis_MC2_155 ----QGAVAGALINTGQDCTAATRAIVARELYDDFVAGVAEVMGKIVVGD
MLBr_02573|M.leprae_Br4923 ----ATAVTGRVQNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGD
MMAR_4246|M.marinum_M ----RMLFESCLLHAGQACILHSRLLLPEAMHDEIVDRLVALARAVRVGD
MUL_0939|M.ulcerans_Agy99 ----RMLFESCLLHAGQACILHSRLLLPEAMHDEIVDRLVALARAVRVGD
MAV_1340|M.avium_104 ----QMLFQSCSLHAGQACILHSRLLLPDSLHDDVVDRLAALARDVKVGD
Mb0791|M.bovis_AF2122/97 ----VSAFSAC-MHAGQGCAITTRLVVPRARYEEAVAIAAATMSSIRPGD
TH_3902|M.thermoresistible__bu ----AHVVGEFTLHAGQGCSLLTRTLVEESIHDELVDWVNKQLAALKVGD
Rv0223c|M.tuberculosis_H37Rv ----PMMVFSGVMNAGQGCVNQTRILAPRSRYDEIVAAVTNFVTALPVGP
MAB_0900c|M.abscessus_ATCC_199 DLYIKQLVAEMTTKAGQKCTAIRRALVPEALVDAAVTATSERLSKVVVGN
. : ** * * : : * : *
Mflv_4860|M.gilvum_PYR-GCK PADPDTDLGPLISAVHREKVAGMVERAPGEGGRVVTGGV----APD--GP
Mvan_1572|M.vanbaalenii_PYR-1 PGDPDTDLGPLITLAHRAKVAGMVERAPGEGGRIVTGGV----APD--GP
MSMEG_1665|M.smegmatis_MC2_155 PHDPDTDLGPLISYAHRDKVAGMVARAPQQGGRVVTGGE----APD--LP
MLBr_02573|M.leprae_Br4923 PTDPDTDVGPLATESGRAQVEQQVDAAAAAGAVIRCGGK----RPD--RP
MMAR_4246|M.marinum_M PADPDVQMGPLISAAQRSRVEDQVARALRDGATLAAGGR----RPATPEA
MUL_0939|M.ulcerans_Agy99 PADPDVQMGPLISAAQRSRVEDQVARALRDGATLAAGGR----RPATPEA
MAV_1340|M.avium_104 PTDPDVQMGPLISAAQRDRVEAHIAGARSDGATLVTGGG----RPAGLDV
Mb0791|M.bovis_AF2122/97 PNDPGTVCGPLISARQRDRVQGYLDLAVAEGGRFACGGA----RPADREV
TH_3902|M.thermoresistible__bu PADPSVMMGPLISEAQRNRVEGYIKAGVDEGAEIAFGGG----RPSHLER
Rv0223c|M.tuberculosis_H37Rv PSDPAAQIGPLISEKQRTRVEGYIAKGIEEGARLVCGGG----RPEGLDN
MAB_0900c|M.abscessus_ATCC_199 PELDSVRMGALASIDQRDEVLRSLKLLQDSATLVFGDPASFTVEGADAAQ
* . *.* : * .* : . . .
Mflv_4860|M.gilvum_PYR-GCK GSFYRPTLIADVA-ESSEVWRDEIFGPVLTVRSFTDDDDAIRQANDTKYG
Mvan_1572|M.vanbaalenii_PYR-1 GSFYRPTLIADVS-EQSEVWRDEIFGPVLAVRSFTDDDDALRQANDTAYG
MSMEG_1665|M.smegmatis_MC2_155 GAFYRPTLIADVD-ESSEVYRDEIFGPVLTVRSFTDDDDALRQANDTEYG
MLBr_02573|M.leprae_Br4923 GWFYPPTVITDIT-KDMALYTDEVFGPVASVYCATNIDDAIEIANATPFG
MMAR_4246|M.marinum_M GFYYEPTILTGVT-PDAHIAQNEVFGPVLSVLRYRDDDDAVAIANNSQYG
MUL_0939|M.ulcerans_Agy99 GFYYEPTILTGVT-PDAHIAQNEVFGPVLSVLRYRDDDDAVAIANNSQYG
MAV_1340|M.avium_104 GYYVEPTILSGVE-PNSTVAQEEVFGPVLSVLRYRDDDDAVAIANNSRYG
Mb0791|M.bovis_AF2122/97 GFYIEPTVIAGLT-NDARVAREEIFGPVLTVIAHDGDDDAVRIANDSPYG
TH_3902|M.thermoresistible__bu GYFVEPTLFTGVR-NEMKIAQEEIFGPVGVVIPFTSEHEAVRLANDSDFG
Rv0223c|M.tuberculosis_H37Rv GFFIQPTVFADVD-NKMTIAQEEIFGPVLAIIPYDTEEDAIAIANDSVYG
MAB_0900c|M.abscessus_ATCC_199 GAFLPPLLLRAEDPTAAAVHEVEAFGPVSTVIGYRDLDDAVELAARGQGS
* : * :: : * **** : .:*: * .
Mflv_4860|M.gilvum_PYR-GCK LAASAWTRD--VYRAQRASREIHAGCVWINDHIPIISE---------MPH
Mvan_1572|M.vanbaalenii_PYR-1 LAASAWTRD--VYRAQRASREIHAGCVWINDHIPIISE---------MPH
MSMEG_1665|M.smegmatis_MC2_155 LAASAWTRD--VYRAQRASREINAGCVWINDHIPIISE---------MPH
MLBr_02573|M.leprae_Br4923 LGSNAWTQN--ESEQRHFIDNIVAGQVFINGMTVSYPE---------LPF
MMAR_4246|M.marinum_M LSGAVWGGD--ADRAVRVARRIRTGQIAVNG--FGPGD---------APF
MUL_0939|M.ulcerans_Agy99 LSGAVWGGD--ADRAVRVARRIRTGQIAVNG--FGPGD---------APF
MAV_1340|M.avium_104 LSGAVWGDD--VDRAVGVARRIRTGQIAVNG--ISPAG---------APF
Mb0791|M.bovis_AF2122/97 LSGTVYGAD--PQRAARIASRLRVGTVNVNGGVWYCAD---------APF
TH_3902|M.thermoresistible__bu LGGGVWSTD--VERALRVAQRIRTGMVAINGGGGSMGKT--------GPF
Rv0223c|M.tuberculosis_H37Rv LAGSVWTTD--VPKGIKISQQIRTGTYGINWYAFDPGS----------PF
MAB_0900c|M.abscessus_ATCC_199 LVGSLVTHDPEVARKVTIGIAAHHGRILVLDRDDAKESTGHGSPLPTLVH
* . : . * : .
Mflv_4860|M.gilvum_PYR-GCK GGFGASGFGKDMS-QYSLEEYLSIKHVMSDITSAVDKDWHRTIFTKR---
Mvan_1572|M.vanbaalenii_PYR-1 GGFGASGFGKDMS-QYSLEEYLSIKHVMSDITGVADKTWHRTIFTKR---
MSMEG_1665|M.smegmatis_MC2_155 GGVGASGFGKDMS-DYSFEEYLTIKHVMSDITGVAEKEWHRTIFNKR---
MLBr_02573|M.leprae_Br4923 GGIKRSGYGRELS-THGIREFCNIKTVWIA--------------------
MMAR_4246|M.marinum_M GGFKQSGLGREGGGIAGLHQYMEPKAIGMPG-------------------
MUL_0939|M.ulcerans_Agy99 GGFKQSGLGREGGGIAGLHQYMEPKAIGMPG-------------------
MAV_1340|M.avium_104 GGFRLSGLGREGGGIGGLHQYMEPMAIGVPA-------------------
Mb0791|M.bovis_AF2122/97 GGYKQSGIGREMG-LLGFEEYLEAKLIATAAN------------------
TH_3902|M.thermoresistible__bu GGFKASGLGREWG-EWGLDEFLEMKQIIWPIAGG----------------
Rv0223c|M.tuberculosis_H37Rv GGYKNSGIGRENG-PEGVEHFTQQKSVLLPMGYTVA--------------
MAB_0900c|M.abscessus_ATCC_199 GGPGRAGGGEELGGIRGVLHFMQRTAIQASPDMLTAITGRWTTGSARTAT
** :* *.: . .. .: :
Mflv_4860|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1572|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1665|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02573|M.leprae_Br4923 --------------------------------------------------
MMAR_4246|M.marinum_M --------------------------------------------------
MUL_0939|M.ulcerans_Agy99 --------------------------------------------------
MAV_1340|M.avium_104 --------------------------------------------------
Mb0791|M.bovis_AF2122/97 --------------------------------------------------
TH_3902|M.thermoresistible__bu --------------------------------------------------
Rv0223c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0900c|M.abscessus_ATCC_199 EVHPFRKHLEELQIGDTIVGGPRVVSLEDIDHFAEFTGDTFYAHTDPDAA
Mflv_4860|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1572|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1665|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02573|M.leprae_Br4923 --------------------------------------------------
MMAR_4246|M.marinum_M --------------------------------------------------
MUL_0939|M.ulcerans_Agy99 --------------------------------------------------
MAV_1340|M.avium_104 --------------------------------------------------
Mb0791|M.bovis_AF2122/97 --------------------------------------------------
TH_3902|M.thermoresistible__bu --------------------------------------------------
Rv0223c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0900c|M.abscessus_ATCC_199 ARNPLFGGIVAHGYLVVSLAAGLFVEPNPGPVLANFGVDGLRFLTPVKPG
Mflv_4860|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1572|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1665|M.smegmatis_MC2_155 --------------------------------------------------
MLBr_02573|M.leprae_Br4923 --------------------------------------------------
MMAR_4246|M.marinum_M --------------------------------------------------
MUL_0939|M.ulcerans_Agy99 --------------------------------------------------
MAV_1340|M.avium_104 --------------------------------------------------
Mb0791|M.bovis_AF2122/97 --------------------------------------------------
TH_3902|M.thermoresistible__bu --------------------------------------------------
Rv0223c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0900c|M.abscessus_ATCC_199 DALTVTLTAKQLTPRLSADYGEVRWDAVVTNQNDDPVATYDVLTLVAKKE
Mflv_4860|M.gilvum_PYR-GCK -
Mvan_1572|M.vanbaalenii_PYR-1 -
MSMEG_1665|M.smegmatis_MC2_155 -
MLBr_02573|M.leprae_Br4923 -
MMAR_4246|M.marinum_M -
MUL_0939|M.ulcerans_Agy99 -
MAV_1340|M.avium_104 -
Mb0791|M.bovis_AF2122/97 -
TH_3902|M.thermoresistible__bu -
Rv0223c|M.tuberculosis_H37Rv -
MAB_0900c|M.abscessus_ATCC_199 S