For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAERALPESLVDTYQLYIDGRWVEPENGRYDDVSPSTGMVIATAPDASVSQVGDAVDAARRAFDAGPWPA MSHQERGHCLAQLGAALQDYADAYFTLSQVEWGCTANERRLQIDASASAARRAGHLASTLGDQPIQSASG AAVLCHEPLGVVSILTPWNFPHSLNVMKLSRALAGGNTVVLKPSPLTPLAGLALARIIHEHTDIPPGVVN VVTPGGIEASKVLTVDPRIDMVSFTGSSAVGCEVMAAAAGTMKRVLLECGGKSASIVLDDAECSDELLAR MLFESCLLHAGQACILHSRLLLPEAMHDEIVDRLVALARAVRVGDPADPDVQMGPLISAAQRSRVEDQVA RALRDGATLAAGGRRPATPEAGFYYEPTILTGVTPDAHIAQNEVFGPVLSVLRYRDDDDAVAIANNSQYG LSGAVWGGDADRAVRVARRIRTGQIAVNGFGPGDAPFGGFKQSGLGREGGGIAGLHQYMEPKAIGMPG
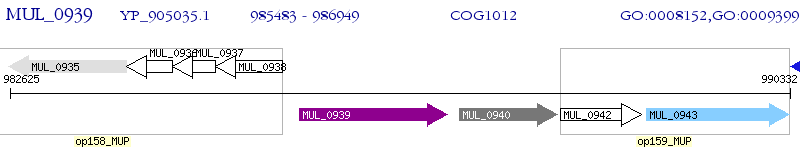
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0939 | aldA_1 | - | 100% (488) | NAD-dependent aldehyde dehydrogenase, AldA_1 |
| M. ulcerans Agy99 | MUL_0477 | aldA | e-101 | 44.14% (478) | NAD-dependent aldehyde dehydrogenase, AldA |
| M. ulcerans Agy99 | MUL_1122 | - | 1e-88 | 39.46% (479) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0791 | aldA | 1e-110 | 46.17% (483) | aldehyde dehydrogenase NAD dependant AldA |
| M. gilvum PYR-GCK | Mflv_2502 | - | 1e-106 | 48.74% (478) | aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0768 | aldA | 1e-110 | 46.17% (483) | aldehyde dehydrogenase NAD dependent AldA |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 1e-54 | 32.21% (444) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1218 | - | 1e-103 | 43.75% (480) | aldehyde dehydrogenase AldA |
| M. marinum M | MMAR_4246 | aldA_1 | 0.0 | 99.18% (488) | NAD-dependent aldehyde dehydrogenase, AldA_1 |
| M. avium 104 | MAV_1340 | - | 0.0 | 72.20% (482) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5859 | - | 1e-105 | 44.47% (479) | aldehyde dehydrogenase (NAD) family protein |
| M. thermoresistible (build 8) | TH_4242 | - | 0.0 | 66.11% (481) | PUTATIVE aldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4156 | - | 1e-101 | 47.17% (477) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0939|M.ulcerans_Agy99 ---MAERALPESLVDT------YQLYIDGRWVEPENGRYDDVSPSTGMVI
MMAR_4246|M.marinum_M ---MAERALPESLVDT------YQLYIDGRWVEPENGRYDDVSPSTGMVI
MAV_1340|M.avium_104 ---MAD---PPALVQT------YQLYIDGQWVEPQDGRYDDLSPCTEAVI
TH_4242|M.thermoresistible__bu ---MTR---SHALVET------YGHYINGEWVDPDSGRYEDVDPATGETF
Mb0791|M.bovis_AF2122/97 ---MALWGDGIS-----------ALLIDGKLSDGRAGTFPTVNPATEEVL
Rv0768|M.tuberculosis_H37Rv ---MALWGDGIS-----------ALLIDGKLSDGRAGTFPTVNPATEEVL
MAB_1218|M.abscessus_ATCC_1997 ---MTVLSDGDS-----------ELFIDGKLLPGGAGRFPTVNPATEEVL
MSMEG_5859|M.smegmatis_MC2_155 ---MPLLADRQS-----------QLLIDGKLVAGRKGVFDTVNPATEEVL
Mflv_2502|M.gilvum_PYR-GCK MQETVTIPDATSGTETADRRAARRLLIGGQLVDTER-TFASLNPATGDVL
Mvan_4156|M.vanbaalenii_PYR-1 MQETATISDERSGAGQAAQRAERHLLIGGQLLETER-TFPSLNPATGEVL
MLBr_02573|M.leprae_Br4923 ------------------------------------MSIATINPATGETV
:.*.* ..
MUL_0939|M.ulcerans_Agy99 ATAPDASVSQVGDAVDAARRAFDAGPWPAMSHQERGHCLAQLGAALQDYA
MMAR_4246|M.marinum_M ATAPDASVSQVGDAVDAARRAFDAGPWPAMSHQERGHCLAQLGAALQDYA
MAV_1340|M.avium_104 ATAPDASVAQVDDAIAAARRAFDSGPWGRISAEQRARCLNQLGEALTKHA
TH_4242|M.thermoresistible__bu AAAPDASTAQVDRAIAAAREAFDSGRWTGLTPEERAKCLQQLGAALLEHA
Mb0791|M.bovis_AF2122/97 GVAADADAEDMGRAIEAARRAFDSTDWSRNT-ELRVRCVRQLRDAMQQHV
Rv0768|M.tuberculosis_H37Rv GVAADADAEDMGRAIEAARRAFDSTDWSRNT-ELRVRCVRQLRDAMQQHV
MAB_1218|M.abscessus_ATCC_1997 GTAADADSDDMSDAIEAARRAFDDSGWSRDV-ALRVHCIRQLRDGMKTHI
MSMEG_5859|M.smegmatis_MC2_155 GVAADATAEDMGDAIEAARRAFDDTDWSTDT-ELRVRCIRQLRDALREHV
Mflv_2502|M.gilvum_PYR-GCK GYAPDASVADAEAAVAAARAAFDTGDWATDT-ELRIRCLEQLHAALVEHR
Mvan_4156|M.vanbaalenii_PYR-1 GYAPDATVADAEAAVAAARRAFDAGVWATDT-ELRIRCLEQFHAALVEHR
MLBr_02573|M.leprae_Br4923 KTFTPTTQDEVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEA
. : : *: * ** : * : :
MUL_0939|M.ulcerans_Agy99 DAYFTLSQVEWG-----CTANERRLQIDASASAARRAGHLASTLGDQPIQ
MMAR_4246|M.marinum_M DAYFALSQVEWG-----CTANERRLQIDASASAARRAGHLAGTLGDQPIQ
MAV_1340|M.avium_104 DDFFALSQTEWG-----CIGNERIMQIDGAGFMSMHAAQLATQLVDQEVA
TH_4242|M.thermoresistible__bu DDFFALAQLEWG-----CSANERMFHVEGPAVMTTHAGELALEPVEQPID
Mb0791|M.bovis_AF2122/97 EELRELTISEVGAPRMLTASAQLEGPVGDLSFAADTAESYPWKQDLGEAS
Rv0768|M.tuberculosis_H37Rv EELRELTISEVGAPRMLTASAQLEGPVGDLSFAADTAESYPWKQDLGEAS
MAB_1218|M.abscessus_ATCC_1997 EELRELTIAEVGAPRMLTAGAQLEGPVDDLQFAADTAESFDWTSDLGVAS
MSMEG_5859|M.smegmatis_MC2_155 EELRDLTIAEVGAPRMLTSGAQLEGPIDDLAFSADTAENYPWRTDLGHAT
Mflv_2502|M.gilvum_PYR-GCK DELAALTIAEVGATPALCQGAQLDQPIEIVRYYADLLKTYPLTEDLGNIE
Mvan_4156|M.vanbaalenii_PYR-1 DELAALTVAEVGATPALCQGAQLDQPIEIVRYYADLLKTYPLTEDLGNIE
MLBr_02573|M.leprae_Br4923 NQSAALMTLEMG-KTLQSAKDEAMKCAKGFRYYAEKAETLLADEPAEAAA
: * * * : :
MUL_0939|M.ulcerans_Agy99 SASG--AAVLCHEPLGVVSILTPWNFPHSLNVMKLSRALAGGNTVVLKPS
MMAR_4246|M.marinum_M SASG--AAVLCHEPLGVVSILTPWNFPHSLNVMKLSRALAGGNTVVLKPS
MAV_1340|M.avium_104 GISAG-TTLLRHVPLGVVAVLTPWNFPHCLNVMKLNHALAAGNTVVLKPS
TH_4242|M.thermoresistible__bu AWGAGGTTLLRYEPLGVVSVLTPWNFPHTLNAMKIGSALAAGNTVVLKPS
Mb0791|M.bovis_AF2122/97 PLGIATRRTLAREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPA
Rv0768|M.tuberculosis_H37Rv PLGIATRRTLAREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPA
MAB_1218|M.abscessus_ATCC_1997 PMGIKTRRTLAREAVGVVGAVTPWNFPHQINLAKVGPALAAGNTLVLKPA
MSMEG_5859|M.smegmatis_MC2_155 PQGIPTDRVIAREAVGVVGAITPWNFPHQINLAKVGPALAAGNTLVLKPA
Mflv_2502|M.gilvum_PYR-GCK SRGMQHHRWVEKEAGGVVAAIIAYNYPNQLALAKLAPALAAGCTVVLKSA
Mvan_4156|M.vanbaalenii_PYR-1 SRGMQHHRWVEKEAGGVVAAIIAYNYPNQLALAKLAPALAAGCTVVLKSA
MLBr_02573|M.leprae_Br4923 VGAS--KALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHA
. . *** : .:*:* ** .* . :** :
MUL_0939|M.ulcerans_Agy99 PLTPLAGLALARIIHEHTDIPPGVVNVVTPGGIEASKVLTVDPRIDMVSF
MMAR_4246|M.marinum_M PLTPLAGLALARIIHEHTDIPPGVVNVVTPGGIEASKVLAVDPRIDMVSF
MAV_1340|M.avium_104 PLTPLAGLALARLIDEHTDIPPGVVNVVTPSGVEAAKLLTTDPRIDMVSF
TH_4242|M.thermoresistible__bu PLTPLAGFALARMIDEHTDIPPGVVNVVTPSGLDGSKMLTTDPRIDMVSF
Mb0791|M.bovis_AF2122/97 PDTPWCAAALGEIIVEHTDFPPGVVNIVTSSSHALGALLAKDPRVDMISF
Rv0768|M.tuberculosis_H37Rv PDTPWCAAALGEIIVEHTDFPPGVVNIVTSSSHALGALLAKDPRVDMISF
MAB_1218|M.abscessus_ATCC_1997 PDTPWCAAVLGRIIAEHTDFPPGVVNIVTSSDHRIGAQLSTDPRVDMVSF
MSMEG_5859|M.smegmatis_MC2_155 PDTPWAAAVLGQIITEHTDFPPGVINIVTSSDHSVGALLSKDPRVDMVSF
Mflv_2502|M.gilvum_PYR-GCK PDTPLITLALGELIAEHTDIPPGVVNVLSGADPQVGAALTTSPDVDMVTF
Mvan_4156|M.vanbaalenii_PYR-1 PDTPLVTLALGELIAKHTDIPAGVVNVLSGADPAVGAALTTSPDVDMVTF
MLBr_02573|M.leprae_Br4923 SNVPQCALYLADVIARG-GFPEDCFQTLLISANGVEAILR-DPRVAAATL
. .* *. :* . .:* . .: : . * .* : ::
MUL_0939|M.ulcerans_Agy99 TGSSAVGCEVMAAAAGTMKRVLLECGGKSASIVLDD-AECSDELLARMLF
MMAR_4246|M.marinum_M TGSSAVGREVMAAAAGTMKRVLLECGGKSASIVLDD-AECSDELLARMLF
MAV_1340|M.avium_104 TGSSAVGRQVMSAAGDTIKRILLELGGKSASIVLDD-ANVTDEMLEQMLF
TH_4242|M.thermoresistible__bu TGSSAVGREVMAGAAGGMKRILLECGGKSASIVLDD-LEVTDELLETILF
Mb0791|M.bovis_AF2122/97 TGSTATGRAVMADAAATIKKVFLELGGKSAFVVLDD---ADLAAASAVSA
Rv0768|M.tuberculosis_H37Rv TGSTATGRAVMADAAATIKKVFLELGGKSAFVVLDD---ADLAAASAVSA
MAB_1218|M.abscessus_ATCC_1997 TGSTATGRAVMTDAAATVKKVFLELGGKSAFIVLDDLDEAALGSACSVAA
MSMEG_5859|M.smegmatis_MC2_155 TGSTATGRAVMTDAALTIKKVFLELGGKSAFLVLDD---ADLTGACSMAA
Mflv_2502|M.gilvum_PYR-GCK TGSTPTGRAIMAAASGTLKKVFLELGGKSAAIVLDD---ADFSTAALFSA
Mvan_4156|M.vanbaalenii_PYR-1 TGSTPTGRAIMAAASGTLKKVFLELGGKSAAIVLDD---ADFNTAAMFSA
MLBr_02573|M.leprae_Br4923 TGSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPS---ADLDAAVATAV
*** ..* : : *. :* .** **... :*: .
MUL_0939|M.ulcerans_Agy99 ESCLLHAGQACILHSRLLLPEAMHDEIVDRLVALARAVRVGDPADPDVQM
MMAR_4246|M.marinum_M ESCLLHAGQACILHSRLLLPEAMHDEIVDRLVALARAVRVGDPADPDVQM
MAV_1340|M.avium_104 QSCSLHAGQACILHSRLLLPDSLHDDVVDRLAALARDVKVGDPTDPDVQM
TH_4242|M.thermoresistible__bu ECLTMHAGQACILNSRLVVPEAMHDDVVARLGELARKVVVGNPVDPAVQM
Mb0791|M.bovis_AF2122/97 FSACMHAGQGCAITTRLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVC
Rv0768|M.tuberculosis_H37Rv FSACMHAGQGCAITTRLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVC
MAB_1218|M.abscessus_ATCC_1997 FTAAMHAGQGCAITTRLVVPRERYDDAVAAAAATMGSLKPGDPAKPGTIC
MSMEG_5859|M.smegmatis_MC2_155 FTVAMHAGQGCAITTRLVVPRARYDEAVEIAAGTLGSIKPGDPTSKRTVC
Mflv_2502|M.gilvum_PYR-GCK FSMVTHAGQGCALTSRLLVPRKHKDEIVELVKNNFGLVRLGDPADPSTYM
Mvan_4156|M.vanbaalenii_PYR-1 FSMVTHAGQGCALTSRLLVPKKHKDELVELVKNNFGLVRYGDPTAAGTYM
MLBr_02573|M.leprae_Br4923 TGRVQNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDV
: **.* .*:: :: * : *:* .
MUL_0939|M.ulcerans_Agy99 GPLISAAQRSRVEDQVARALRDGATLAAGGRRPATPEAGFYYEPTILTGV
MMAR_4246|M.marinum_M GPLISAAQRSRVEDQVARALRDGATLAAGGRRPATPEAGFYYEPTILTGV
MAV_1340|M.avium_104 GPLISAAQRDRVEAHIAGARSDGATLVTGGGRPAGLDVGYYVEPTILSGV
TH_4242|M.thermoresistible__bu GPLISRAHLERVEGFVERAVADGATVVAGGSRPAHLDRGFFYEPTILTDV
Mb0791|M.bovis_AF2122/97 GPLISARQRDRVQGYLDLAVAEGGRFACGGARPADREVGFYIEPTVIAGL
Rv0768|M.tuberculosis_H37Rv GPLISARQRDRVQGYLDLAVAEGGRFACGGARPADREVGFYIEPTVIAGL
MAB_1218|M.abscessus_ATCC_1997 GPVISARQRDRVQSYLDLAVAEGGSFACGGGRPEGKDRGFFIEPTVVAGL
MSMEG_5859|M.smegmatis_MC2_155 GPVISARQRDRVQSYLDLAIKEGGRFACGGGRPADMDKGFWIEPTVIAGL
Mflv_2502|M.gilvum_PYR-GCK GPLISEKQRDKVDGMVKRAVEAGATLVTGGEK---VDPGYFYTPTLLADV
Mvan_4156|M.vanbaalenii_PYR-1 GPLISEKQRDKVDGMVKRAVEAGATLVTGGEK---VDPGYFYTPTLLADV
MLBr_02573|M.leprae_Br4923 GPLATESGRAQVEQQVDAAAAAGAVIRCGGKRPD--RPGWFYPPTVITDI
**: : :*: : * *. . ** : *:: **:::.:
MUL_0939|M.ulcerans_Agy99 TPDAHIAQNEVFGPVLSVLRYRDDDDAVAIANNSQYGLSGAVWGGDADRA
MMAR_4246|M.marinum_M TPDAHIAQNEVFGPVLSVLRYRDDDDAVAIANNSQYGLSGAVWGGDADRA
MAV_1340|M.avium_104 EPNSTVAQEEVFGPVLSVLRYRDDDDAVAIANNSRYGLSGAVWGDDVDRA
TH_4242|M.thermoresistible__bu APDDFIAQEEVFGPVLTVLRCRDDDEAVDIANNSRYGLGGAVYAADVDRA
Mb0791|M.bovis_AF2122/97 TNDARVAREEIFGPVLTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRA
Rv0768|M.tuberculosis_H37Rv TNDARVAREEIFGPVLTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRA
MAB_1218|M.abscessus_ATCC_1997 TNAARVAREEIFGPVLVVIAHDGDEDAVRIANDSPYGLSGTVFGDDDERS
MSMEG_5859|M.smegmatis_MC2_155 DNNARVAREEIFGPVLTVIAHDGDDDAVRIANDSPYGLSGTVFSGDDARA
Mflv_2502|M.gilvum_PYR-GCK DPDSEIAQEEVFGPVLTVIAYEDDDDAVRIANNSIYGLSGAVFGSQD-RA
Mvan_4156|M.vanbaalenii_PYR-1 DPDSEIAQEEVFGPVLVVIAYEDDDDAVRIANNSIYGLSGAVFGSQD-RA
MLBr_02573|M.leprae_Br4923 TKDMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQ
: :*:**** * . ::*: *** : :**.. .: : .
MUL_0939|M.ulcerans_Agy99 VRVARRIRTGQIAVNG--FGPGDAPFGGFKQSGLGREGGGIAGLHQYMEP
MMAR_4246|M.marinum_M VRVARRIRTGQIAVNG--FGPGDAPFGGFKQSGLGREGGGIAGLHQYMEP
MAV_1340|M.avium_104 VGVARRIRTGQIAVNG--ISPAGAPFGGFRLSGLGREGGGIGGLHQYMEP
TH_4242|M.thermoresistible__bu LGVARRIRSGQVSING--CIAGDAPFGGFGQSGIGREGG-LLGLRSYMEP
Mb0791|M.bovis_AF2122/97 ARIASRLRVGTVNVNGGVWYCADAPFGGYKQSGIGREMG-LLGFEEYLEA
Rv0768|M.tuberculosis_H37Rv ARIASRLRVGTVNVNGGVWYCADAPFGGYKQSGIGREMG-LLGFEEYLEA
MAB_1218|M.abscessus_ATCC_1997 ARVASRLRVGTVNVNGGVWYSADAPFGGYKQSGNGREMG-VAGFEEYLET
MSMEG_5859|M.smegmatis_MC2_155 QAVASRMRVGTVNVNGGIWYSADAPFGGYKQSGVGREMG-LAGFEEYTEI
Mflv_2502|M.gilvum_PYR-GCK LGVARRIRTGTFSINGGNYFSPDSPFGGYKQSGIGREMG-RAGLEEFLES
Mvan_4156|M.vanbaalenii_PYR-1 LAVARRIRTGTFSINGGNYFAPDSPFGGYKQSGIGREMG-RAGLEEFLES
MLBr_02573|M.leprae_Br4923 RHFIDNIVAGQVFINGMTVSYPELPFGGIKRSGYGRELS-THGIREFCNI
. .: * . :** **** ** *** . *:..: :
MUL_0939|M.ulcerans_Agy99 KAIGMPG-------
MMAR_4246|M.marinum_M KAIGMPG-------
MAV_1340|M.avium_104 MAIGVPA-------
TH_4242|M.thermoresistible__bu KAIGVPA-------
Mb0791|M.bovis_AF2122/97 KLIATAAN------
Rv0768|M.tuberculosis_H37Rv KLIATAAN------
MAB_1218|M.abscessus_ATCC_1997 KVIATAVK------
MSMEG_5859|M.smegmatis_MC2_155 KLIATLAG------
Mflv_2502|M.gilvum_PYR-GCK KTFATVAG------
Mvan_4156|M.vanbaalenii_PYR-1 KTYAVVVPEAGGQA
MLBr_02573|M.leprae_Br4923 KTVWIA--------