For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KTAVTGAAGFIGTNLVNLLVDNGHEVIAIDRVVPASPETREAVTWVSGDVLDPDSMTKALDGVEVVYHLV AVITLKHEDELCWRINTEGARTVAQAALTVGARRMVHCSSIDSYSNSVATIDEKSPRSAAADLPVYQRSK WGGEVAVRETIASGLDAVICNPTGVYGPVDLPNLSRINQLLFDSARGRLPGMVNSQYDLVDARDVAAGLY LAGEKGRTGENYLLGGHMGSLLQVCRLAARRAGKRGPRFAVPMGLLNAAIPVIEPIGKLLKNDALSRAAF NKLTVSPAVDITKARTELGYEPRSTETTVNEFVDFLLGSGRL*
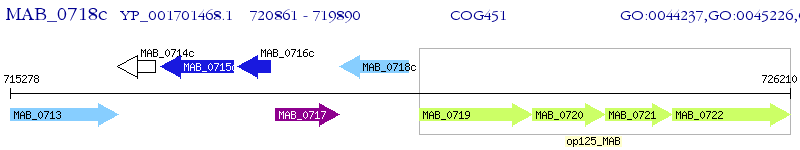
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0718c | - | - | 100% (323) | NAD-dependent epimerase/dehydratase |
| M. abscessus ATCC 19977 | MAB_1682 | - | 2e-23 | 29.31% (331) | NAD-dependent epimerase/dehydratase |
| M. abscessus ATCC 19977 | MAB_2971c | - | 2e-20 | 30.18% (328) | putative oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0144 | - | 4e-21 | 28.35% (328) | putative oxidoreductase |
| M. gilvum PYR-GCK | Mflv_2375 | - | 1e-24 | 27.83% (309) | NAD-dependent epimerase/dehydratase |
| M. tuberculosis H37Rv | Rv0139 | - | 1e-21 | 28.66% (328) | oxidoreductase |
| M. leprae Br4923 | MLBr_01942 | - | 1e-15 | 23.72% (333) | putative cholesterol dehydrogenase |
| M. marinum M | MMAR_0164 | - | 4e-29 | 32.32% (328) | nucleoside-diphosphate-sugar epimerase |
| M. avium 104 | MAV_5157 | - | 1e-24 | 30.58% (327) | 3-beta hydroxysteroid dehydrogenase/isomerase family protein |
| M. smegmatis MC2 155 | MSMEG_6474 | - | 2e-24 | 30.18% (328) | nucleoside-diphosphate-sugar epimerase |
| M. thermoresistible (build 8) | TH_2143 | - | 6e-26 | 27.08% (325) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4932 | - | 2e-29 | 32.01% (328) | nucleoside-diphosphate-sugar epimerases |
| M. vanbaalenii PYR-1 | Mvan_5705 | - | 2e-25 | 30.18% (328) | NAD-dependent epimerase/dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2375|M.gilvum_PYR-GCK ------------MAGTKLVIGASGFLGSHVTRQLVADGD---ADVRVLIR
TH_2143|M.thermoresistible__bu ------------VAKKKLVIGASGFLGSHVTRQLVREG----HDVRVMVR
Mb0144|M.bovis_AF2122/97 -----------MN-APKLVIGANGFLGSHVTRQLVADCAPQKGEVRAMVR
Rv0139|M.tuberculosis_H37Rv -----------MN-APKLVIGANGFLGSHVTRQLVADCAPQKGEVRAMVR
MAV_5157|M.avium_104 -----------MSGKPKLVIGANGFLGSHVTRQLVADGA----QVRAMVR
MSMEG_6474|M.smegmatis_MC2_155 -------------MSTSLVIGANGFLGSHVLRQLVADNT-DGSEIRAMVR
Mvan_5705|M.vanbaalenii_PYR-1 --------------MTALVIGANGYLGSHVTRRLVAD----GQDVRVMVR
MMAR_0164|M.marinum_M ------------MAPRILVTGATGYLGSTILEALVRAG----ERATILVQ
MUL_4932|M.ulcerans_Agy99 ------------MAPRILVTGATGYLGSTILEALVRAD----ERATILVQ
MAB_0718c|M.abscessus_ATCC_199 --------------MKTAVTGAAGFIGTNLVNLLVDNG-------HEVIA
MLBr_01942|M.leprae_Br4923 MLRGVGNASLTTELGRVLVTGGSGFVGANLVTALLERG-------YQVRS
* *. *::*: : *: :
Mflv_2375|M.gilvum_PYR-GCK RTSSTRGIDGLP--VEVRYGDIFDADALRTAMSGCDVVYYCVVDARPWLR
TH_2143|M.thermoresistible__bu TTSMTRGIDDLD--VERCHGDVFDDAALRAAMTGVDDVYYCVVDARMWLR
Mb0144|M.bovis_AF2122/97 PAANTRSIDDLP--LTRFHGDVFDTATVAEAMAGCDDVYYCVVDTRAWLR
Rv0139|M.tuberculosis_H37Rv PAANTRSIDDLP--LTRFHGDVFDTATVAEAMAGCDDVYYCVVDTRAWLR
MAV_5157|M.avium_104 AGANTRGIDDLS--LTRFHGDVFDTAVLSEAMDGVDDVYYCVVDTRAWVR
MSMEG_6474|M.smegmatis_MC2_155 PGANTVGIDDLD--VTRFTGDIFDTEVLRAAMTGCDVVYYCVVDTRGWLR
Mvan_5705|M.vanbaalenii_PYR-1 EGANTIGIDDLT--VTRFVGDIWDDDVLREAMTGCDVVYYCVVDTRGWLR
MMAR_0164|M.marinum_M PGDPHVMSPELRSNVDVVRGDITDAQSVDEAMRGIARVYHLAGIASPNSR
MUL_4932|M.ulcerans_Agy99 PGDPHVMSPELRSNVDVVRGDITDAQSVDEAMRGIARVYHLAGIASPNSR
MAB_0718c|M.abscessus_ATCC_199 IDRVVPASPETREAVTWVSGDVLDPDSMTKALDGVEVVYHLVAVIT-LKH
MLBr_01942|M.leprae_Br4923 FDRAPMPLPQHP-QLEVLQGDITDATVCTTAMDSIDTVFHTAAIIELMGG
: **: * *: . *:: .
Mflv_2375|M.gilvum_PYR-GCK DPR------PMWRTNVDGLRTVLDIAAD----AGLTRFVFTSSIGTIGLR
TH_2143|M.thermoresistible__bu DPA------PLFRTNVEGLRHVLDAAVE----ADLNRFLFTSTTGTMAIN
Mb0144|M.bovis_AF2122/97 DPS------PLFRTNVAGLRNVLDVAT----DASLRRFVFTSSYATVGRR
Rv0139|M.tuberculosis_H37Rv DPS------PLFRTNVAGLRNVLDVAT----DASLRRFVFTSSYATVGRR
MAV_5157|M.avium_104 DTS------PLFRTNVEGLRNVLDVAVT---QPELRKFIFTSTYATVGRR
MSMEG_6474|M.smegmatis_MC2_155 HPA------PLFRTNVEGTRHVLDVALEPG--IGLRKFVYTSSYVTVGRR
Mvan_5705|M.vanbaalenii_PYR-1 DPA------PLFRTNVEGTRNVLNIAVEPAVAAGLRKFVFTSSYVTVGRR
MMAR_0164|M.marinum_M LAN------QIWRTNVLGAYHVAQSAWR----HGVQRLVHVSSTAAIGYP
MUL_4932|M.ulcerans_Agy99 LAN------QIWRTNVLGAYHVAQSAWR----HGVQRLVHASSTAAIGYP
MAB_0718c|M.abscessus_ATCC_199 EDE------LCWRINTEGARTVAQAALT----VGARRMVHCSSIDSYSN-
MLBr_01942|M.leprae_Br4923 ASVTDEYRQRSYTVNVGGTENLLRAGQK----SGVKRFVYTASNSVVMGG
: *. * : . :::. ::
Mflv_2375|M.gilvum_PYR-GCK AGGPADERTEHNWLDRGGEYIRSRVVAEEMVLRDSARNR---LPGVAMCV
TH_2143|M.thermoresistible__bu PYRPVTEDDPHNWT-EGGAYIEARVAAEQLVLRYARERG---LPAVALCI
Mb0144|M.bovis_AF2122/97 RGHVATEEDRVDTR-KVTPYVRSRVAAEDLVLQYAHDAG---LPAVAMCV
Rv0139|M.tuberculosis_H37Rv RGHVATEEDRVDTR-KVTPYVRSRVAAEDLVLQYAHDAG---LPAVAMCV
MAV_5157|M.avium_104 RGHVATEDDVIGTR-GLSDYVKSRVQAENLVMRYVAEAG---LPAVAMCV
MSMEG_6474|M.smegmatis_MC2_155 RGCVATEDDVIDLR-GVTPYVRSRVQAEELVLRYATERG---LPAVAMCV
Mvan_5705|M.vanbaalenii_PYR-1 RGRVATEADVIADR-GLTPYVRSRVQAENLVLEYARTRG---LPAVAMCV
MMAR_0164|M.marinum_M PNGVIADEDFDPRDSVLDNVYSATKRAGEQLVLDFVDRG---LDVVVVNP
MUL_4932|M.ulcerans_Agy99 PNGVIADEDFDPRDSVLDNVYSATKRAGEQLVLDFVDRG---LDVVVVNP
MAB_0718c|M.abscessus_ATCC_199 -SVATIDEKSPRSAAADLPVYQRSKWGGEVAVRETIASG---LDAVICNP
MLBr_01942|M.leprae_Br4923 TPITGGDETMPYTKRFNDLYTETKVVAEKFVLSQNGVPDGETMLTCSIRP
: . . : :
Mflv_2375|M.gilvum_PYR-GCK ANTYGPGDFQPTPHGAMLAAAVRGKLPFYIRGYEAEAVGIEDAARAMILA
TH_2143|M.thermoresistible__bu STTYGPGDWQPTPHGRLLAMVATGRFPFYL-GWSAEVVGIEDAARAMLLA
Mb0144|M.bovis_AF2122/97 STTYGGGDWGRTPHGAFIAGAVFGRLPFTMRGIRLEAVGVDDAARALILA
Rv0139|M.tuberculosis_H37Rv STTYGGGDWGRTPHGAFIAGAVFGRLPFTMRGIRLEAVGVDDAARALILA
MAV_5157|M.avium_104 STTYGSGDWGRTPHGAFIAGAVFGKLPFTMEGIQLEVVGVTDAAKAMVLA
MSMEG_6474|M.smegmatis_MC2_155 STTYGSGDWGRTPHGAVIAGAAFGKLPFVMSGIDLEAVGVDDAARALILA
Mvan_5705|M.vanbaalenii_PYR-1 STTYGAGDWGRTPHGAIIAGAAFGKLPFVLGGIELEAVGVEDAAHALLLA
MMAR_0164|M.marinum_M AAVFAPGFGPPRSWQGLLVAARKGLLRVVPPGGTAVCS-ARDFAAGVTAA
MUL_4932|M.ulcerans_Agy99 AAVFAPGFGPPRSWQGLLVSARKGLLRVVPPGGTAVCS-ARDFAAGVTAA
MAB_0718c|M.abscessus_ATCC_199 TGVYGPVDLPNLSRINQLLFDSARGRLPGMVNSQYDLVDARDVAAGLYLA
MLBr_01942|M.leprae_Br4923 SGIWGRGDQTMFRKAFESVVSGHVKVLIGSKNAKLDNSYVHNLVHGLILA
: :. . : . .: *
Mflv_2375|M.gilvum_PYR-GCK GERGRIG------ERYIVSDRFISTREIHEIGCAAVGMRPPRFGVPIRAM
TH_2143|M.thermoresistible__bu AEQGRVG------ERYIISDRYLHTREVHAIAAEAVGRRAPWFGLPLPVL
Mb0144|M.bovis_AF2122/97 AERGHNG------ERYLISERMMPLQEVVRIAADEAGVPPPRWSISVPVL
Rv0139|M.tuberculosis_H37Rv AERGRNG------ERYLISERMMPLQEVVRIAADEAGVPPPRWSISVPVL
MAV_5157|M.avium_104 ADRGRVG------ERYLISERMIALKEVVRIAADEAGVPPPRRSISVPTL
MSMEG_6474|M.smegmatis_MC2_155 AESGRPG------ERYLISEKLISNAEVARIAAEAAGVAPPRRSIPLPVS
Mvan_5705|M.vanbaalenii_PYR-1 AEKGRVG------ERYLISEKMISNADVVRIAAEAAGVPAPSRTMPLPLS
MMAR_0164|M.marinum_M MNKGQPG------RRYILSTDNLSYRQIAELLVRAVGRSHQVRVAPMRPF
MUL_4932|M.ulcerans_Agy99 MNKGQPG------RRYILSTDNLSYRQIAELLVRAVGRSHQVRVAPMRPF
MAB_0718c|M.abscessus_ATCC_199 GEKGRTG------ENYLLGGHMGSLLQVCRLAARRAGKRGPRFAVPMGLL
MLBr_01942|M.leprae_Br4923 AEHLVPGGTAPGQAYFINDGEPINFFDFMGPIIKACGENWPRVRISGRLV
: * :: . :. * .
Mflv_2375|M.gilvum_PYR-GCK AAAAHLGAAVARVRN---TDTMLTPLNIRLMHIMTPLDHSKAVRELGWHP
TH_2143|M.thermoresistible__bu SVAARGNDLAARLLR---RDLMFAHVGLKMAELMSPLDHSKAERELGFKP
Mb0144|M.bovis_AF2122/97 YALGALGSLRARLTG---KDTELSLASVRMMRSEADVDHGKAVRELGWQP
Rv0139|M.tuberculosis_H37Rv YALGALGSLRARLTG---KDTELSLASVRMMRSEADVDHGKAVRELGWQP
MAV_5157|M.avium_104 YALGALGDLRARLTG---KDAELSLASVRMMRAEAPVDHSKAVRELGWQP
MSMEG_6474|M.smegmatis_MC2_155 YLLAAIGTVKGKLRG---TDERLSLRSLRLMRAECELDHSKAVRELGWQP
Mvan_5705|M.vanbaalenii_PYR-1 YAMAALGSLKGRLKG---TDERLSLDSLRLMRAEAPVDCGKAKRELGWQP
MMAR_0164|M.marinum_M RALGRGKRIASDFSSRAHFDDPLVPENIDLMARKVYYDPTRAVRELGIPK
MUL_4932|M.ulcerans_Agy99 RALGMGKRIASDFSSRAHFDDPLVPENIDLMARKVYYDPTRAVRELGIPK
MAB_0718c|M.abscessus_ATCC_199 NAAIPVIEPIGKLLK----NDALSRAAFNKLTVSPAVDITKARTELGYEP
MLBr_01942|M.leprae_Br4923 RNVMAVWQRLHFGFG--LPKPPMEPLAVERVYLDNYFSIEKAHKELGYRP
. : . . :* ***
Mflv_2375|M.gilvum_PYR-GCK R-PLADSIVSAAHFFTDRRRSAAPEGTR------------
TH_2143|M.thermoresistible__bu E-PVEDSIAKAARFFLEQHR--------------------
Mb0144|M.bovis_AF2122/97 R-PVEESIREAARFWAAMRTVGKDPAAS------------
Rv0139|M.tuberculosis_H37Rv R-PVEESIREAARFWAAMRTVGKDPAAS------------
MAV_5157|M.avium_104 R-PVEESIREAARFWAAMRNAKGKSATPR-----------
MSMEG_6474|M.smegmatis_MC2_155 R-PVEESIAEAAKFWVGLRAAKQARRGG------------
Mvan_5705|M.vanbaalenii_PYR-1 R-PVEESIREAAKFWVGLRAAKRAGKAAG-----------
MMAR_0164|M.marinum_M V-SVAELITEFVG---------------------------
MUL_4932|M.ulcerans_Agy99 V-SVAELITEFVG---------------------------
MAB_0718c|M.abscessus_ATCC_199 R-STETTVNEFVDFLLGSGRL-------------------
MLBr_01942|M.leprae_Br4923 LFTTEQAMAECLPYYTELFEQIKVAAKPHLASIAATPRPE
. : .