For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLRGVGNASLTTELGRVLVTGGSGFVGANLVTALLERGYQVRSFDRAPMPLPQHPQLEVLQGDITDATVC TTAMDSIDTVFHTAAIIELMGGASVTDEYRQRSYTVNVGGTENLLRAGQKSGVKRFVYTASNSVVMGGTP ITGGDETMPYTKRFNDLYTETKVVAEKFVLSQNGVPDGETMLTCSIRPSGIWGRGDQTMFRKAFESVVSG HVKVLIGSKNAKLDNSYVHNLVHGLILAAEHLVPGGTAPGQAYFINDGEPINFFDFMGPIIKACGENWPR VRISGRLVRNVMAVWQRLHFGFGLPKPPMEPLAVERVYLDNYFSIEKAHKELGYRPLFTTEQAMAECLPY YTELFEQIKVAAKPHLASIAATPRPE
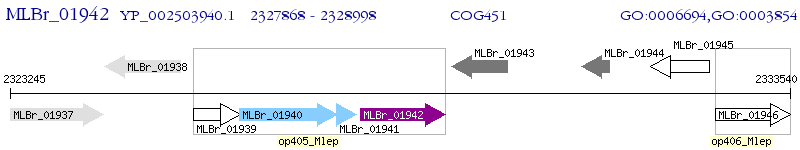
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01942 | - | - | 100% (376) | putative cholesterol dehydrogenase |
| M. leprae Br4923 | MLBr_00204 | rmlB2 | 3e-11 | 27.63% (257) | putative sugar-nucleotide dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1136c | - | 1e-165 | 75.47% (375) | cholesterol dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2075 | - | 1e-153 | 71.39% (367) | 3-beta hydroxysteroid dehydrogenase/isomerase |
| M. tuberculosis H37Rv | Rv1106c | - | 1e-165 | 75.47% (375) | cholesterol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4385 | - | 6e-16 | 27.54% (334) | putative oxidoreductase |
| M. marinum M | MMAR_4359 | - | 1e-164 | 75.76% (363) | cholesterol dehydrogenase |
| M. avium 104 | MAV_1225 | - | 1e-165 | 75.82% (368) | 3-beta hydroxysteroid dehydrogenase/isomerase family protein |
| M. smegmatis MC2 155 | MSMEG_5228 | - | 1e-153 | 73.11% (357) | 3-beta hydroxysteroid dehydrogenase/isomerase family protein |
| M. thermoresistible (build 8) | TH_1114 | - | 1e-159 | 74.37% (355) | PROBABLE CHOLESTEROL DEHYDROGENASE |
| M. ulcerans Agy99 | MUL_0172 | - | 1e-165 | 75.76% (363) | cholesterol dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4635 | - | 1e-157 | 74.18% (368) | 3-beta hydroxysteroid dehydrogenase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2075|M.gilvum_PYR-GCK --------MLRTELGRVLVTGGSGFVGANLVTELLDRGLHVRSFDRVASA
Mvan_4635|M.vanbaalenii_PYR-1 ---MGDATTPTTELGRVLVTGGSGFVGANLVTELLDRGLQVRSFDRVPSP
MSMEG_5228|M.smegmatis_MC2_155 ------MADSTTDLGRILVTGGSGFVGANLVTELLDRGYAVRSFDRVPSP
TH_1114|M.thermoresistible__bu ----MGDPTLRTDLGRVLVTGGSGFVGTNLVTTLLERGHHVRSCDRVGSP
Mb1136c|M.bovis_AF2122/97 MLRRMGDASLTTELGRVLVTGGAGFVGANLVTTLLDRGHWVRSFDRAPSL
Rv1106c|M.tuberculosis_H37Rv MLRRMGDASLTTELGRVLVTGGAGFVGANLVTTLLDRGHWVRSFDRAPSL
MAV_1225|M.avium_104 MIGPMGDPSLTAELGRVLVTGGSGFVGANLVTTLLERGYQVRSFDRAPSP
MMAR_4359|M.marinum_M MLRPMGDASLTTELGRVLVTGGSGFVGSNLVTTLLDRGHHVRSFDRAPSP
MUL_0172|M.ulcerans_Agy99 MLRPMGDASLTTELGRVLVTGGSGFVGSNLVTTLLDRGHHVRSFDRAPSP
MLBr_01942|M.leprae_Br4923 MLRGVGNASLTTELGRVLVTGGSGFVGANLVTALLERGYQVRSFDRAPMP
MAB_4385|M.abscessus_ATCC_1997 ---------MTDMTQRAFVTGANGFVGRSLSLRLRTLGWQVSGVD-----
* :***. **** .* * * * . *
Mflv_2075|M.gilvum_PYR-GCK LPAHARLEIVEGDITDADDVAAAVEG----VDTVFHTAAIIDLMGGASVS
Mvan_4635|M.vanbaalenii_PYR-1 LPDHPGLEVVQGDITDVDDVARAVGTGADKADTVFHTAAIIDLMGGASVT
MSMEG_5228|M.smegmatis_MC2_155 LPAHAGLEVATGDICDLDNVTNAVAG----VDTVFHTAAIIDLMGGASVT
TH_1114|M.thermoresistible__bu LPDHPRLEVIDGDICDADAVAAAVDG----IDTVFHTAAVIDLMGGAGVT
Mb1136c|M.bovis_AF2122/97 LPAHPQLEVLQGDITDADVCAAAVDG----IDTIFHTAAIIELMGGASVT
Rv1106c|M.tuberculosis_H37Rv LPAHPQLEVLQGDITDADVCAAAVDG----IDTIFHTAAIIELMGGASVT
MAV_1225|M.avium_104 LPAHPHLEVLQGDITDAGVCAAAVEG----IDTIFHTAAIIDLMGGASVT
MMAR_4359|M.marinum_M LPPHPQLEVLQGDITDAATCATAVDG----VDTVFHTAAIIELMGGASVT
MUL_0172|M.ulcerans_Agy99 LPPHPQLEVLQGDITDAATCATAVDG----VDTVFHTAAIIELMGGASVT
MLBr_01942|M.leprae_Br4923 LPQHPQLEVLQGDITDATVCTTAMDS----IDTVFHTAAIIELMGGASVT
MAB_4385|M.abscessus_ATCC_1997 LVADPAHNIQGGDINTAGPWMTEAAA----ADAIFHTAAVVSNIADS---
* .. :: *** *::*****::. :..:
Mflv_2075|M.gilvum_PYR-GCK EEYRQRSFAVNVTGTQNLVHAAQKAGVTRFVYTASNSVVMGGQRIAGGDE
Mvan_4635|M.vanbaalenii_PYR-1 EEYRQRSFAVNVTGTKNLVHAAQKAGVQRFVYTASNSVVMGGQRIAGGDE
MSMEG_5228|M.smegmatis_MC2_155 AEYRQRSFAVNVGGTENLVRAAQSAGVKRFVYTASNSVVMGGQHIVHGDE
TH_1114|M.thermoresistible__bu DEYRERSFAVNVHGTERLVRAAQQAGVRRFVYTASNSVVMGGQPIADGDE
Mb1136c|M.bovis_AF2122/97 DEYRQRSFAVNVGGTENLLHAGQRAGVQRFVYTSSNSVVMGGQNIAGGDE
Rv1106c|M.tuberculosis_H37Rv DEYRQRSFAVNVGGTENLLHAGQRAGVQRFVYTSSNSVVMGGQNIAGGDE
MAV_1225|M.avium_104 EEYRQRSFAVNVGGTENLVRAGQAAGVQRFVYTSSNSVVMGGQNIVGGDE
MMAR_4359|M.marinum_M DEYRKRSFSVNVGGTENLVREGQQAGVKRFVYTSSNSVVMGGQIIAGGDE
MUL_0172|M.ulcerans_Agy99 DEYRKRSFSVNVGGTENLVRAGQQAGVKRFVYTSSNSVVMGGQIIAGGDE
MLBr_01942|M.leprae_Br4923 DEYRQRSYTVNVGGTENLLRAGQKSGVKRFVYTASNSVVMGGTPITGGDE
MAB_4385|M.abscessus_ATCC_1997 ----DTAWRVNVLGTKKVIDATAPGAVFVHLSSVRAFSDLDFPDMVTEEH
. :: *** **:.:: ..* .: : :. :. :.
Mflv_2075|M.gilvum_PYR-GCK TLPYTERFNDLYTETKVVAEKFVLSQNGV---SGLLTCSIRPSGIWGRGD
Mvan_4635|M.vanbaalenii_PYR-1 TLPYTERFNDLYTETKVVAEKFVLSQNGV---SGMLTCSIRPSGIWGRGD
MSMEG_5228|M.smegmatis_MC2_155 TLPYTERFNDLYTETKVVAEKFVLGRNGV---AGMLTCSIRPSGIWGRGD
TH_1114|M.thermoresistible__bu TLPYTDRFNDLYTETKVIAERFVLGQNGI---EGMLTCSIRPSGIWGPGD
Mb1136c|M.bovis_AF2122/97 TLPYTDRFNDLYTETKVVAERFVLAQNGV---DGMLTCAIRPSGIWGNGD
Rv1106c|M.tuberculosis_H37Rv TLPYTDRFNDLYTETKVVAERFVLAQNGV---DGMLTCAIRPSGIWGNGD
MAV_1225|M.avium_104 TLPYTDRFNDLYTETKVLAERFVLGQNGV---DGMLTCAIRPSGIWGRGD
MMAR_4359|M.marinum_M TLPYTNRFNDLYTETKVVAEKFVLSQNGN---NEMLTCAIRPSGIWGTGD
MUL_0172|M.ulcerans_Agy99 TLPYTNRFNDLYTETKVVAEKFVLSQNGN---NEMLTCAIRPSGIWGTGD
MLBr_01942|M.leprae_Br4923 TMPYTKRFNDLYTETKVVAEKFVLSQNGVPDGETMLTCSIRPSGIWGRGD
MAB_4385|M.abscessus_ATCC_1997 PVRTDG---NAYVDTKVASEQVVLQAHGAG---RIDARIARPGDIYGPGS
.: : *.:*** :*:.** :* : : **..*:* *.
Mflv_2075|M.gilvum_PYR-GCK QTMFRKVFESVLAGHVKVLVGNKETKLDNSYVHNLVHGFILAAEHLVEGG
Mvan_4635|M.vanbaalenii_PYR-1 QTMFRKVFESVLAGHVKVLVGNKNVKLDNSYVHNLVHGFILAAQHLVDGG
MSMEG_5228|M.smegmatis_MC2_155 QTMFRKVFESVLAGHVKVLVGRKSTLLDNSYVHNLVHGFILAAEHLTPNG
TH_1114|M.thermoresistible__bu QTMFRKLFESVHAGHVKVLIGSRHARLDNSYVHNLVHGFILAAEHLVPGG
Mb1136c|M.bovis_AF2122/97 QTMFRKLFESVLKGHVKVLVGRKSARLDNSYVHNLIHGFILAAAHLVPDG
Rv1106c|M.tuberculosis_H37Rv QTMFRKLFESVLKGHVKVLVGRKSARLDNSYVHNLIHGFILAAAHLVPDG
MAV_1225|M.avium_104 QTMFRKLFESVIAGHVKVLIGRKSARLDNSYVHNLIHGFILAAQHLTPGG
MMAR_4359|M.marinum_M QLMFRKLFESVIKGHVKVLVGPKSALLDNSYVDNLIHGFILAAQHLVPGG
MUL_0172|M.ulcerans_Agy99 QLMFRKLFESVIKGHVKVLVGPKSVLLDNSYVDNLIHGFMLAAQHLVPGG
MLBr_01942|M.leprae_Br4923 QTMFRKAFESVVSGHVKVLIGSKNAKLDNSYVHNLVHGLILAAEHLVPGG
MAB_4385|M.abscessus_ATCC_1997 -RPWTILPVEMIRSRQFVLPAMGRGVFSPIYIDDLVEGVVALGS----GG
: .: .: ** . :. *:.:*:.*.: . .*
Mflv_2075|M.gilvum_PYR-GCK SAPGQAYFINDGEPINMFEFARPVVQACGEPFPKFRVPGRLVWFAMTIWQ
Mvan_4635|M.vanbaalenii_PYR-1 TAPGQAYFINDGEPINMFEFARPVMEACGEPWPTFRVSGRLVWFAMTIWQ
MSMEG_5228|M.smegmatis_MC2_155 TAPGQAYFINDGEPVNMFEFARPVIEACGRKLPRVRVPGRAVHAAMSGWQ
TH_1114|M.thermoresistible__bu TAPGQAYFINDGEPINMFEFARPVVTACGQRWPTLRVSGPLVRAVMSLWQ
Mb1136c|M.bovis_AF2122/97 TAPGQAYFINDAEPINMFEFARPVLEACGQRWPKMRISGPAVRWVMTGWQ
Rv1106c|M.tuberculosis_H37Rv TAPGQAYFINDAEPINMFEFARPVLEACGQRWPKMRISGPAVRWVMTGWQ
MAV_1225|M.avium_104 TAPGQAYFINDAEPINMFEFARPVVEACGVNWPRVRVNGPIVRAAMTGWQ
MMAR_4359|M.marinum_M SAPGQAYFINDAEPINMFEFARPVVEACGEKWPRVRVSGPMVHRAMTGWQ
MUL_0172|M.ulcerans_Agy99 SAPGQAYFINDAEPINMFDFARPVVEACGEKWPRVRVSGPMVHRAMTGWQ
MLBr_01942|M.leprae_Br4923 TAPGQAYFINDGEPINFFDFMGPIIKACGENWPRVRISGRLVRNVMAVWQ
MAB_4385|M.abscessus_ATCC_1997 AASGQVFTISGGVGVTCAQFFGHYHRMLDRRGPLVLPTAAAVGLATVKAR
:*.**.: *... :. :* . * . . * . :
Mflv_2075|M.gilvum_PYR-GCK FLHFKFGLPKPLLEPLAVERLYLDNYFSIAKAQRDLGYQPLFTTEQALAQ
Mvan_4635|M.vanbaalenii_PYR-1 FLHFRFGLPKPLLEPLAVERLYLDNYFSIAKAERDLGYRPLYTTEQALEH
MSMEG_5228|M.smegmatis_MC2_155 RLHFRFGIPEPLLEPLAVERLYLNNYFSIAKATRDLGYRPLFTTEQARVD
TH_1114|M.thermoresistible__bu RLHFRFGLPKPPLEPLAVERLYLDNYFSIDKARRELGYRPRYTTEQALQE
Mb1136c|M.bovis_AF2122/97 RLHFRFGFPAPLLEPLAVERLYLDNYFSIAKARRDLGYEPLFTTQQALTE
Rv1106c|M.tuberculosis_H37Rv RLHFRFGFPAPLLEPLAVERLYLDNYFSIAKARRDLGYEPLFTTQQALTE
MAV_1225|M.avium_104 RLHFRFGIPAPLLEPLAVERLYLDNFFSIAKASRDLGYQPLFTTEQAMSE
MMAR_4359|M.marinum_M RLHFRFGLPAPLLEPLAVERLYLDNYFSVDKARRDLGYEPKFTTEQALKE
MUL_0172|M.ulcerans_Agy99 RLHFRFGLPAPLLEPLAVERLYLDNYFSVDKARRDLGYEPKFTTEQALKE
MLBr_01942|M.leprae_Br4923 RLHFGFGLPKPPMEPLAVERVYLDNYFSIEKAHKELGYRPLFTTEQAMAE
MAB_4385|M.abscessus_ATCC_1997 VLALSG--KATEANAISVRYLARTGSYSIAKAQRILGFEPRFSLDAGMRA
* : . :.::*. : . :*: ** : **:.* :: : .
Mflv_2075|M.gilvum_PYR-GCK CIPYYVELFDRMKREAGNPVVQAAAPAPPKG
Mvan_4635|M.vanbaalenii_PYR-1 CIPYYVDLFGRMKAEAAQPAVQAAAPVPPAG
MSMEG_5228|M.smegmatis_MC2_155 CLPYYVDLFKQMEAQAR-PA-----------
TH_1114|M.thermoresistible__bu CLPYYVDLFHRMKAEPATV------------
Mb1136c|M.bovis_AF2122/97 CLPYYVSLFEQMKNEARAEKTAATVKP----
Rv1106c|M.tuberculosis_H37Rv CLPYYVSLFEQMKNEARAEKTAATVKP----
MAV_1225|M.avium_104 CLPYYVGMFEQMKRQALAGKASA--------
MMAR_4359|M.marinum_M CLPYYVDLFRQMKSEVKAGGR----------
MUL_0172|M.ulcerans_Agy99 CLPYYVDLFRQMKSEVKAGGR----------
MLBr_01942|M.leprae_Br4923 CLPYYTELFEQIKVAAKPHLASIAATPRPE-
MAB_4385|M.abscessus_ATCC_1997 TEIWLRDQGYLD-------------------
: