For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDQPARPLRADAARNRARVLEVAYQTFAEEGLGVPIDEIARRAGVGAGTVYRHFPAKEDLYRAIAEDRMG SVIDKGRELLAAVDPGEALFEFLRSMMDWGGTDQGLVDALSGAGIDVQALIPHIEESFYAVLGDLLRAAQ QAGTVRSDVGVHEVKALLVGCQAMQSYNKDVSRQVTDVVFDGLRPRP
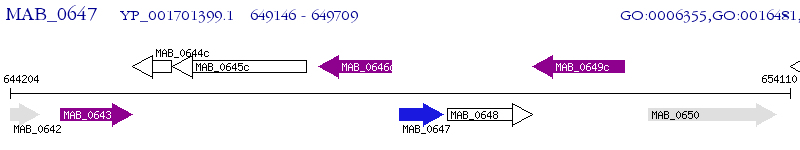
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0647 | - | - | 100% (187) | transcriptional regulator TetR |
| M. abscessus ATCC 19977 | MAB_1873c | - | 4e-20 | 33.15% (184) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_4384 | - | 3e-17 | 32.63% (190) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0068c | - | 2e-64 | 67.22% (180) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0142 | - | 2e-57 | 62.78% (180) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0067c | - | 2e-64 | 67.22% (180) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00316 | - | 2e-64 | 65.59% (186) | putative TetR/AcrR-family transcriptional regulator |
| M. marinum M | MMAR_0489 | - | 2e-08 | 29.81% (161) | TetR family transcriptional regulator |
| M. avium 104 | MAV_2463 | - | 4e-23 | 36.41% (195) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_0878 | - | 1e-63 | 64.64% (181) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_1262 | - | 4e-09 | 36.90% (84) | transcriptional regulator, TetR family protein |
| M. ulcerans Agy99 | MUL_1152 | - | 2e-08 | 29.81% (161) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0763 | - | 6e-59 | 64.64% (181) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0489|M.marinum_M ----------MPTVT--WARVDPARRAAIVEAAEAEFGAHGFSQGSLNVI
MUL_1152|M.ulcerans_Agy99 ----------MPTVT--WARVDPARRAAIVEAAEAEFGAHGFSQGSLNVI
MAV_2463|M.avium_104 ----------MTAVEDRPLRSDAARNVERILRAAREVYAELGPDAPVEAV
Mb0068c|M.bovis_AF2122/97 -----------MAPTDRRVRADAARNRARVLEVAYQTFAADGLSVPVDEI
Rv0067c|M.tuberculosis_H37Rv -----------MAPTDRRVRADAARNRARVLEVAYQTFAADGLSVPVDEI
MLBr_00316|M.leprae_Br4923 MSVPRSGLSVQMAQPARPLRADAARNRARILGVAYEAFATEGLSVPIDEI
MSMEG_0878|M.smegmatis_MC2_155 -----------MS-----MRADAARNRARVLEVAYEAFAQDGLAVPIDEI
Mflv_0142|M.gilvum_PYR-GCK -----------MT---RPLRADAARNRARVLEVAYDTFAAEGLTVPIDEI
Mvan_0763|M.vanbaalenii_PYR-1 -----------MT---RPLRADAARNRARVLEVAYDTFAAEGLGVPIDEI
MAB_0647|M.abscessus_ATCC_1997 -----------MDQPARPLRADAARNRARVLEVAYQTFAEEGLGVPIDEI
TH_1262|M.thermoresistible__bu ---------MRTHGWGGSAPASDEEAIARILEAAKRTIEERGAEFSIADV
. . : . .: :
MMAR_0489|M.marinum_M ARRARVAKGSLFQYFADKRDLYAYIADIASQRVRTHIEGLIR-ELDSSRP
MUL_1152|M.ulcerans_Agy99 ARRARVAKGSLFQYFADKRDLYAYIADIANQRVRTHIEGLIR-ELDSSRP
MAV_2463|M.avium_104 ARRAGVGERTLYRRFPAKADLVRAALDQS------IAEDLTP-VIDSARH
Mb0068c|M.bovis_AF2122/97 ARRAGVGAGTVYRHFPTKEALFQAVIADRMHRIIDKGHALLK-SKHPGDA
Rv0067c|M.tuberculosis_H37Rv ARRAGVGAGTVYRHFPTKEALFQAVIADRMHRIIDKGHALLK-SKHPGDA
MLBr_00316|M.leprae_Br4923 ARRAGVGAGTVYRHFPTKEALCAAVIGDRMPHLVDDGYVLLK-SAGPGEA
MSMEG_0878|M.smegmatis_MC2_155 ARRAGVGAGTVYRHFPSKEDLYRAVVAERIREIVDQGQALLA-DGDPGEA
Mflv_0142|M.gilvum_PYR-GCK ARRAGVGAGTVYRHFPTKEALVVAVAEDRVRRIAERARTLLA-VEGPGQA
Mvan_0763|M.vanbaalenii_PYR-1 ARRAGVGAGTVYRHFPTKQALVVAVAEDRVRRIVDHARTLLA-AEGPGEA
MAB_0647|M.abscessus_ATCC_1997 ARRAGVGAGTVYRHFPAKEDLYRAIAEDRMGSVIDKGRELLA-AVDPGEA
TH_1262|M.thermoresistible__bu ARAVGVTRQTVYRYFPSTDALLVAAAMHAAGDFLDRLAVHLQGITDPVEA
** . * :::: *. . * .
MMAR_0489|M.marinum_M FFEFLTDLLDGWVAYFAEHPRERALHAAATLEVDTDARISVRSVIHRHYL
MUL_1152|M.ulcerans_Agy99 FFEFLTDLLDGWVAYFAEHPRERALHAAATLEVDTDARISVRSVIHRHYL
MAV_2463|M.avium_104 AKDPLHGLTRLVEAAISLGAREQNLLAAAHRAGSLTSDISVS------LN
Mb0068c|M.bovis_AF2122/97 LFAFLRSMVLQW------GATDRGLVEALAGVG-IEISSAAP-EAEADFL
Rv0067c|M.tuberculosis_H37Rv LFAFLRSMVLQW------GATDRGLVEALAGVG-IEISSAAP-EAEADFL
MLBr_00316|M.leprae_Br4923 LFTYLRSLVLHW------GATDRGLVDALAGAGGIDILGVAP-DAEDAFL
MSMEG_0878|M.smegmatis_MC2_155 LFDYLRLMVLQC------GAADRGLVEALAGAG-IDVKTMVP-DAEDEYM
Mflv_0142|M.gilvum_PYR-GCK LFVFFRDMVRS-------AAVDHGLVDALMNQG-LDLEAVAP-GVEAAFL
Mvan_0763|M.vanbaalenii_PYR-1 LFVFLRDMVRS-------AAADYGLVDALVGYG-LDLEVAAP-GAEAAFL
MAB_0647|M.abscessus_ATCC_1997 LFEFLRS-MMDW------GGTDQGLVDALSGAG-IDVQALIP-HIEESFY
TH_1262|M.thermoresistible__bu VAEAIATTLEWLP-----EDTHIGLLVAPGRPT-AHTESVTSDVAIDFAN
: . * *
MMAR_0489|M.marinum_M EVLRPLVRDALARGDLRADSDTDALLSLLLMIFPHLALAPYMRGLDPILG
MUL_1152|M.ulcerans_Agy99 EVLRPLVRDALARGDLRADSDTDALLSLLLMIFPHLALAPYMRGLDPILG
MAV_2463|M.avium_104 EALGELVREGQRAGRIRADLVADDLPRLVAMLYS--VLSTMDAGSEGWRR
Mb0068c|M.bovis_AF2122/97 DLLTDLLRAAQRAGTVRPDVDVLEVKTLLVGCQAMQSYNAELAAKVTDVA
Rv0067c|M.tuberculosis_H37Rv DLLTDLLRAAQRAGTVRPDVDVLEVKTLLVGCQAMQSYNAELAAKVTDVA
MLBr_00316|M.leprae_Br4923 TILSDLLYAAQYAGTARTDVGVREVKSILVGCQAMEAYNSALAERVTDVV
MSMEG_0878|M.smegmatis_MC2_155 EMLGGLLRAAQRAGTVRGDVTATDVKALLVGCQAMQAYDATAAEHLTGVV
Mflv_0142|M.gilvum_PYR-GCK STLGDLLSAAQGAGTVRPDVDVAVVKALLVVCKVPQVFDSEVTERVTRVI
Mvan_0763|M.vanbaalenii_PYR-1 ATLGELLAAAQRAGTVRADVDVAVIKALLVVCKVPQVYDGEVSERVVRVI
MAB_0647|M.abscessus_ATCC_1997 AVLGDLLRAAQQAGTVRSDVGVHEVKALLVGCQAMQSYNKDVSRQVTDVV
TH_1262|M.thermoresistible__bu RMLRRFDVDWAAVGYSDADLDELAEHLLRVIQSFVIDPGRPPRDGQSLRA
* : * * :
MMAR_0489|M.marinum_M LDEPTPEQPALAVRRLVAVLTAAYSPSLTVEVRKVT
MUL_1152|M.ulcerans_Agy99 LDEPTPEQPALAVRRLVAVLTAAYSPSLTVEVRKVT
MAV_2463|M.avium_104 YVALVVDAISVDERRPLPPAAALRYASQPNSWPL--
Mb0068c|M.bovis_AF2122/97 LDGLRANRK---------------------------
Rv0067c|M.tuberculosis_H37Rv LDGLRANRK---------------------------
MLBr_00316|M.leprae_Br4923 VDGLRAAR----------------------------
MSMEG_0878|M.smegmatis_MC2_155 FDGLRVAPASVPSR----------------------
Mflv_0142|M.gilvum_PYR-GCK EDGLRPLR----------------------------
Mvan_0763|M.vanbaalenii_PYR-1 EDGLRVRSSV--------------------------
MAB_0647|M.abscessus_ATCC_1997 FDGLRPRP----------------------------
TH_1262|M.thermoresistible__bu YLRRWVGAAVTGAR----------------------