For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QFGVLTFVTDEGIGPAELGAALEHRGFESLFLAEHTHIPVNTQSPYPGGGPIPEKYYRTLDPFVALAAAA ATTQSLVLGTGIALIPERDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVIDERLR AIIEIWTQEQAEFHGTYVDFDPIYCWPKPVTKPYPPLYVGGGPANFPRIARLNAGWIAISPSPQRLSGPL QRLRAMAGGDVPVTVCQWGEAAAKDLEGYRHLGVERVLLELPTEPRDPTLRYLDKLQAELARLA*
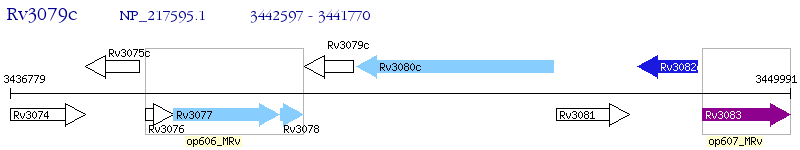
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3079c | - | - | 100% (275) | hypothetical protein Rv3079c |
| M. tuberculosis H37Rv | Rv0953c | - | 6e-50 | 40.57% (281) | oxidoreductase |
| M. tuberculosis H37Rv | Rv2161c | - | 9e-43 | 40.88% (274) | hypothetical protein Rv2161c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3106c | - | 1e-162 | 100.00% (275) | hypothetical protein Mb3106c |
| M. gilvum PYR-GCK | Mflv_1017 | - | 1e-73 | 51.27% (275) | luciferase family protein |
| M. leprae Br4923 | MLBr_00348 | - | 5e-06 | 25.82% (213) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1059c | - | 1e-47 | 36.40% (283) | hypothetical protein MAB_1059c |
| M. marinum M | MMAR_1575 | - | 1e-129 | 81.39% (274) | oxidoreductase |
| M. avium 104 | MAV_1077 | - | 9e-48 | 39.86% (276) | hypothetical protein MAV_1077 |
| M. smegmatis MC2 155 | MSMEG_1996 | - | 1e-70 | 50.94% (267) | N5,N10- methylenetetrahydromethanopterin reductase-related |
| M. thermoresistible (build 8) | TH_2750 | - | 7e-75 | 50.71% (280) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_3458 | - | 1e-129 | 81.02% (274) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5819 | - | 5e-72 | 51.27% (275) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3079c|M.tuberculosis_H37Rv -----------MQFGVLTFVTDEGIGPAELGAALEHRGFESLFLAEHTHI
Mb3106c|M.bovis_AF2122/97 -----------MQFGVLTFVTDEGIGPAELGAALEHRGFESLFLAEHTHI
MMAR_1575|M.marinum_M -----------MRVGVLTFVTDEGIGPAELGAALEERGFESLFLAEHTHI
MUL_3458|M.ulcerans_Agy99 -----------MRVGVLTFVTDEGIGPAELGAALEERGFESLFLAEHTHI
Mflv_1017|M.gilvum_PYR-GCK -----------MKFGISTFPNEDTIDPVSLARAVEERGFAALAVAEHTHI
Mvan_5819|M.vanbaalenii_PYR-1 -----------MKFGIATFVNDTTIDPVALARAVEDRGFGSLAVAEHTHI
MSMEG_1996|M.smegmatis_MC2_155 MHTVINSQGTGMKIGISTFVNDDTIDAVTLARAIEERGFHSLVIAEHSHI
TH_2750|M.thermoresistible__bu -----------MRFGISTFVNDDTIDPVSLATAIEERGFDALTVAEHTHI
MAB_1059c|M.abscessus_ATCC_199 -----------MDYGLVLFTSDRGISPAIAAKAAEDGGFSTFYVPEHTHI
MAV_1077|M.avium_104 ----------------MLFTSDRGISPAAAAKLADDHGFATFYVPEHTHI
MLBr_00348|M.leprae_Br4923 -----------------------MVERANAMKLGLQLGYWAAQPPGNAAE
: . . *: : . ::
Rv3079c|M.tuberculosis_H37Rv PVNTQSPYP--GGGPIP-EKYYRTLDPFVALAAAAATTQSLVLGTGIALI
Mb3106c|M.bovis_AF2122/97 PVNTQSPYP--GGGPIP-EKYYRTLDPFVALAAAAATTQSLVLGTGIALI
MMAR_1575|M.marinum_M PVNNQSPYP--PGGPIP-RKYYRTLDPFVALTAAAVATESLVLGTGIALI
MUL_3458|M.ulcerans_Agy99 PVNNQSPYP--PGGPIP-RKYYRTLDPFVALTAAAVATESLVLGTGIALI
Mflv_1017|M.gilvum_PYR-GCK PASRESAYP--LGGELP-KIYYRTLDPFVTLAAAAAVTSEIELITGIALL
Mvan_5819|M.vanbaalenii_PYR-1 PASRESAYP--LGGELP-SIYYRTLDPFVTLAAAAAVTTSIELVTGIALL
MSMEG_1996|M.smegmatis_MC2_155 PASRESAYP--GGDELP-QKYYRTLDPFVTLAAAAAVTTKIELFTGVALL
TH_2750|M.thermoresistible__bu PASRESPYP--EGGELP-SVYYRTLDPFVTLAAAAAVTSRIQLITGIALL
MAB_1059c|M.abscessus_ATCC_199 PVKREAAHPGTGGAELPDDRYMRTLDPWVSLGAACAVTSKIRLATAVAIP
MAV_1077|M.avium_104 PIKREAAHPTTGDASLPDDRYMRTLDPWVSLGAACAVTSRVRLSTAVALP
MLBr_00348|M.leprae_Br4923 LVAAAEGAG--FDAVFTGEAWG--SDAYTPLAWWGSSTQRVRLGTSVVQL
. :. : *.:..* * : * *.:.
Rv3079c|M.tuberculosis_H37Rv PERDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVI
Mb3106c|M.bovis_AF2122/97 PERDPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVANHGVDPAVRGRVI
MMAR_1575|M.marinum_M PERDPIVTAKEVASLDVVSGGRFCFGVGVGWLREEVANHGVDPAVRGRVT
MUL_3458|M.ulcerans_Agy99 PERDPIVTAKEVASLDVVSGGRFCFGVGVGWLREEVANHGVDPAVRGRVS
Mflv_1017|M.gilvum_PYR-GCK IQRDPIITAKEAASIDVISGGRFVFGVGAGWNIEELRHHGTDPKTRGALL
Mvan_5819|M.vanbaalenii_PYR-1 IQRDPIITAKEAASIDLISNGRFVFGVGAGWNLEELRHHGTDPKTRGALL
MSMEG_1996|M.smegmatis_MC2_155 IQRDVIHTAKEAASVDLISNGRFVFGVGAGWNLEEMRNHGTDPKTRGRLL
TH_2750|M.thermoresistible__bu IQRDPIITAKEAASIDLISDGRFVFGVGAGWNLEEMRHHGTDPRTRGALL
MAB_1059c|M.abscessus_ATCC_199 VEHDPITLAKSIATLDHMSGGRVTVGVGYGWNTDELADHHVPAGRRRTML
MAV_1077|M.avium_104 VEHDPITLAKSIATLDHLSGGRVSLGVGFGWNTDELADHGVPAGRRRTML
MLBr_00348|M.leprae_Br4923 SARTPTACAMAALTLDHLSGGRHILGLGVSGPQVVEGWYGQPFPKPLSRT
: * ::* :* ** .*:* . :
Rv3079c|M.tuberculosis_H37Rv DERLRAIIEIWTQEQ-AEFHGTYVDFDPIYCWPKPVTKPYP---------
Mb3106c|M.bovis_AF2122/97 DERLRAIIEIWTQEQ-AEFHGTYVDFDPIYCWPKPVTKPYP---------
MMAR_1575|M.marinum_M DERLGAMIEIWTQDN-AEYHGKYVDFDPICCWPKPVTKPHP---------
MUL_3458|M.ulcerans_Agy99 DERLGAMIEIWTQDN-AEYHGKYVDFDPICCWPKPVTKPHP---------
Mflv_1017|M.gilvum_PYR-GCK DERIEAIKVLWTQEP-AEYHGRFVDFDASYQRPKPVRKPHP---------
Mvan_5819|M.vanbaalenii_PYR-1 DERIEAIKALWTAEP-AEYHGKYVDFDASYLRPKPVQKPHP---------
MSMEG_1996|M.smegmatis_MC2_155 DEQIEAIKALWTQEP-AEYHGKYVDIESSYMRPKPVQKPHP---------
TH_2750|M.thermoresistible__bu DERIEAVKALWTDEP-AEYHGRFVDFGPSYLRPKPVQKPHP---------
MAB_1059c|M.abscessus_ATCC_199 REYIEAMRRLWEEEE-ASYSGEFVNFGPSWAWPKTVQKHVP---------
MAV_1077|M.avium_104 REYLEAMRALWTQEE-AAYDGEFVKFGPSWAWPKPVQPHIP---------
MLBr_00348|M.leprae_Br4923 REYIDIVRQVWAREAPVVSDGQHYPLPLTGAGATGLGKALKPITHPLRAD
* : : :* : . * . : .. :
Rv3079c|M.tuberculosis_H37Rv -PLYVGGGP-ANFPRIARLNAGWIAISPSPQRLS----------------
Mb3106c|M.bovis_AF2122/97 -PLYVGGGP-ANFPRIARLNAGWIAISPSPQRLS----------------
MMAR_1575|M.marinum_M -PLYIGGGP-ASFGRIARLGAGWIAISPSPQLLA----------------
MUL_3458|M.ulcerans_Agy99 -PLYIGGGP-ASFGRIARLGAGWIAISPSPQLLA----------------
Mflv_1017|M.gilvum_PYR-GCK -PILIGGDSDATVRRIIRHGAGWIANPLPIDHLS----------------
Mvan_5819|M.vanbaalenii_PYR-1 -PILIGGDSDAAVKRIIRHEAGWISNPLPVDTLR----------------
MSMEG_1996|M.smegmatis_MC2_155 -PIYIGGASEATVKRVIRHDAGWISNPLPVDFLS----------------
TH_2750|M.thermoresistible__bu -PIVVGGNSDATVRRIIRHGAGWMSNPHPPEMLK----------------
MAB_1059c|M.abscessus_ATCC_199 -VIVGAGGTEKNFTWIAKNADGWMSTPRDEDIAGKI--------------
MAV_1077|M.avium_104 -VLVGAAGNEKNFKWIARSADGWITTPRDFDIDEPV--------------
MLBr_00348|M.leprae_Br4923 IPIMLGAEGPKNVALAAEICDGWLPIFFSPRMADMYNEWLDEGFARTGAR
: .. . . **:.
Rv3079c|M.tuberculosis_H37Rv -------------GPLQRLRAMAGGD-VPVTVCQWGEAAAK----DLEGY
Mb3106c|M.bovis_AF2122/97 -------------GPLQRLRAMAGGD-VPVTVCQWGEAAAK----DLEGY
MMAR_1575|M.marinum_M -------------GPLEELRSLAGED-VAVTVCLSDETKVS----DLEGY
MUL_3458|M.ulcerans_Agy99 -------------GPLEELRSLAGEE-VAVTVCLSDETKVS----DLEGY
Mflv_1017|M.gilvum_PYR-GCK -------------RRIEQIREGAGGD-VPLCQFG-TPVKPE----YWQAA
Mvan_5819|M.vanbaalenii_PYR-1 -------------RRVDQIRAGARHE-VPLSMFG-TPVKPE----YWQAA
MSMEG_1996|M.smegmatis_MC2_155 -------------KFVTQMRDGLGHE-VPLAQFG-TPADPE----YWGAV
TH_2750|M.thermoresistible__bu -------------RRVQQIRDGAGHD-VLLTTFG-TPVDPG----YWEVL
MAB_1059c|M.abscessus_ATCC_199 -------------VALKEAWKAAGRDGDPLIYVLAGKPDPE----QLAAW
MAV_1077|M.avium_104 -------------KLLQDTWAAAGRDGAPQIVALDFKPDAE----KLARW
MLBr_00348|M.leprae_Br4923 RSREDFEICASAQIVVTEDRAAAFAGIKPFLALYMGGMGAEGTNFHADVY
:
Rv3079c|M.tuberculosis_H37Rv RHLGVERVLLELPTEPRDP------------------TLRYLDKLQAELA
Mb3106c|M.bovis_AF2122/97 RHLGVERVLLELPTEPRDP------------------TLRYLDKLQAELA
MMAR_1575|M.marinum_M LPLGVERVLLGLPTEPRDQ------------------TLRYLDQMESELV
MUL_3458|M.ulcerans_Agy99 LPLGVERVLLGLPTEPRDQ------------------TLRYLDQMESELV
Mflv_1017|M.gilvum_PYR-GCK EELGFGQLNLLLPTKSRDV------------------SLRLLDDYAEQVA
Mvan_5819|M.vanbaalenii_PYR-1 EDLGFGQLNLLLPTKPRDE------------------SLRLLDGYAEQVA
MSMEG_1996|M.smegmatis_MC2_155 EELGFGQIALLLPTLPTDE------------------SLRMLDDYATAVE
TH_2750|M.thermoresistible__bu EELGYQQANLLLPTRPRDE------------------SLRMLDQFAERVA
MAB_1059c|M.abscessus_ATCC_199 RELGVTEAIFGMPDRSEEE------------------VRAYIDRLSGKLA
MAV_1077|M.avium_104 AELGVTEVLFGLPDKSEDE------------------VAAYVERLAGKLA
MLBr_00348|M.leprae_Br4923 RRMGYAEVVDEVTALFRSNQKDKAAEIIPNELVDSTMIVGDVDYVCKQIA
:* . :. . :: :
Rv3079c|M.tuberculosis_H37Rv RLA------------------------
Mb3106c|M.bovis_AF2122/97 RLA------------------------
MMAR_1575|M.marinum_M QLG------------------------
MUL_3458|M.ulcerans_Agy99 QLG------------------------
Mflv_1017|M.gilvum_PYR-GCK RYIG-----------------------
Mvan_5819|M.vanbaalenii_PYR-1 RY-------------------------
MSMEG_1996|M.smegmatis_MC2_155 KYRG-----------------------
TH_2750|M.thermoresistible__bu GYRG-----------------------
MAB_1059c|M.abscessus_ATCC_199 AFA------------------------
MAV_1077|M.avium_104 ALV------------------------
MLBr_00348|M.leprae_Br4923 AWSAAGVTMMMVSASSVEQVRDLAGLF