For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGFGISTTSADGYVSPDVLARAVEERGFEWLLFDDHSYFPVDTPDSIDAEFRARLPLIRDLPVVLAYAAS ATERLIVGSGVALLPQRDVLHTAKAWATVATLSGGRTVLGVGVGWNLGEVRNHGIDPAKRGAVLDEQLAA LHALWTQEQAEFHGEHVDFGPVVARPRPARPLPVHVGGPSRAAQARALRVGDGWLPYAGTSTPDDVRRAC GWFAEQGRADLPVTVSGVPGDHQLAGEYLGAGAERVLFDVHPLNEADTLRALDRLTEVVARLT*
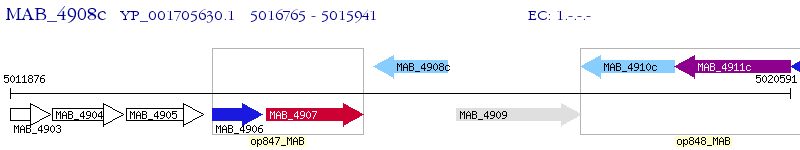
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4908c | - | - | 100% (274) | putative luciferase-like oxidoreductase |
| M. abscessus ATCC 19977 | MAB_1059c | - | 2e-26 | 30.04% (283) | hypothetical protein MAB_1059c |
| M. abscessus ATCC 19977 | MAB_0366 | - | 2e-21 | 34.50% (200) | hypothetical protein MAB_0366 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3106c | - | 6e-44 | 40.00% (275) | hypothetical protein Mb3106c |
| M. gilvum PYR-GCK | Mflv_1017 | - | 4e-44 | 40.49% (284) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3079c | - | 6e-44 | 40.00% (275) | hypothetical protein Rv3079c |
| M. leprae Br4923 | MLBr_00269 | - | 7e-06 | 28.89% (135) | putative F420-dependent glucose-6-phosphate dehydrogenase |
| M. marinum M | MMAR_3559 | - | 9e-47 | 41.07% (280) | hypothetical protein MMAR_3559 |
| M. avium 104 | MAV_1077 | - | 8e-30 | 32.37% (278) | hypothetical protein MAV_1077 |
| M. smegmatis MC2 155 | MSMEG_6703 | - | 1e-105 | 71.16% (267) | N5,N10- methylenetetrahydromethanopterin reductase-related |
| M. thermoresistible (build 8) | TH_2750 | - | 7e-45 | 40.51% (274) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_2792 | - | 1e-43 | 41.04% (251) | hypothetical protein MUL_2792 |
| M. vanbaalenii PYR-1 | Mvan_5819 | - | 4e-44 | 39.64% (275) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4908c|M.abscessus_ATCC_199 MTG---FGISTTSADGYVSP---DVLARAVEERGFEWLLFDDHSYFPVD-
MSMEG_6703|M.smegmatis_MC2_155 MTDAPRFGISTSSADGGIAP---DELARAVEERRFDWLLFDDHSYFPAD-
Mflv_1017|M.gilvum_PYR-GCK ----MKFGISTFPNEDTIDP---VSLARAVEERGFAALAVAEHTHIPASR
Mvan_5819|M.vanbaalenii_PYR-1 ----MKFGIATFVNDTTIDP---VALARAVEDRGFGSLAVAEHTHIPASR
TH_2750|M.thermoresistible__bu ----MRFGISTFVNDDTIDP---VSLATAIEERGFDALTVAEHTHIPASR
MMAR_3559|M.marinum_M ----MKFGISTFVNDDSIDP---VSLARAIEERGFSSLVIAEHTHIPASR
MUL_2792|M.ulcerans_Agy99 ----MKFGISTFVNDDSIDP---VSLARAIEERGFSSLVIAEHTHIPASR
Mb3106c|M.bovis_AF2122/97 ----MQFGVLTFVTDEGIGP---AELGAALEHRGFESLFLAEHTHIPVNT
Rv3079c|M.tuberculosis_H37Rv ----MQFGVLTFVTDEGIGP---AELGAALEHRGFESLFLAEHTHIPVNT
MAV_1077|M.avium_104 ---------MLFTSDRGISP---AAAAKLADDHGFATFYVPEHTHIPIKR
MLBr_00269|M.leprae_Br4923 ---MAELRLGYKASAEQFAPRELVELGVAAEAHGMDSATVSDH-FQPWRH
. * . : : : . :* . *
MAB_4908c|M.abscessus_ATCC_199 -TPDSI---DAEFR-ARLPLIRDLPVVLAYAASATERLIVGSGVALLPQR
MSMEG_6703|M.smegmatis_MC2_155 -TPDAV---DPVFR-ARLPLIRDLFVVLAYAASATSALTIGSGVALLPQR
Mflv_1017|M.gilvum_PYR-GCK ESAYPL---GGELP-KIYYRTLDPFVTLAAAAAVTSEIELITGIALLIQR
Mvan_5819|M.vanbaalenii_PYR-1 ESAYPL---GGELP-SIYYRTLDPFVTLAAAAAVTTSIELVTGIALLIQR
TH_2750|M.thermoresistible__bu ESPYPE---GGELP-SVYYRTLDPFVTLAAAAAVTSRIQLITGIALLIQR
MMAR_3559|M.marinum_M ETPYPG---GGELP-GVYYRTLDPFVTLAAAAAVTSKIELFTGIALLIQR
MUL_2792|M.ulcerans_Agy99 ETPYPG---GDELP-GVYYRTLDPFVTLAAAAAVTSKIELFTGIALLIQR
Mb3106c|M.bovis_AF2122/97 QSPYPG---GGPIP-EKYYRTLDPFVALAAAAATTQSLVLGTGIALIPER
Rv3079c|M.tuberculosis_H37Rv QSPYPG---GGPIP-EKYYRTLDPFVALAAAAATTQSLVLGTGIALIPER
MAV_1077|M.avium_104 EAAHPTT-GDASLPDDRYMRTLDPWVSLGAACAVTSRVRLSTAVALPVEH
MLBr_00269|M.leprae_Br4923 QGGHASFS----------------LSWMTAVGERTNRILLGTSVLTPTFR
. : . * : : :.: :
MAB_4908c|M.abscessus_ATCC_199 -DVLHTAKAWATVATLSGGRTVLGVGVGWNLGEVRN--HGIDPAKRG--A
MSMEG_6703|M.smegmatis_MC2_155 -DTLHTAKAVASLATLSGNRVVLGVGVGWNLNDVRN--HGADPRRRG--A
Mflv_1017|M.gilvum_PYR-GCK -DPIITAKEAASIDVISGGRFVFGVGAGWNIEELRH--HGTDPKTRG--A
Mvan_5819|M.vanbaalenii_PYR-1 -DPIITAKEAASIDLISNGRFVFGVGAGWNLEELRH--HGTDPKTRG--A
TH_2750|M.thermoresistible__bu -DPIITAKEAASIDLISDGRFVFGVGAGWNLEEMRH--HGTDPRTRG--A
MMAR_3559|M.marinum_M -DPITTAKEAASIDLISGGRFVFGVGAGWNIEELRD--HGTDPKTRG--A
MUL_2792|M.ulcerans_Agy99 -DPITTAKEAASIDLISGGRFVFGVGAGWNIEELRD--HGTDPKTRG--A
Mb3106c|M.bovis_AF2122/97 -DPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVAN--HGVDPAVRG--R
Rv3079c|M.tuberculosis_H37Rv -DPIVTAKEVASLDLVSQGRFRFGVGVGWLREEVAN--HGVDPAVRG--R
MAV_1077|M.avium_104 -DPITLAKSIATLDHLSGGRVSLGVGFGWNTDELAD--HGVPAGRRR--T
MLBr_00269|M.leprae_Br4923 YNPAVIGQAFATMGCLYPNRVFLGVGTGEALNEVATGYQGAWPEFKERFA
: .: *:: : .* :*** * :: :* . :
MAB_4908c|M.abscessus_ATCC_199 VLDEQLAALHALWTQEQAEFHGEHVDFGPVVARPRPARP--LPVHVGGPS
MSMEG_6703|M.smegmatis_MC2_155 KLDEQLSALRALWSDDEAEFSGTQVRFGPVVARPRPSAP--VPILVGGAS
Mflv_1017|M.gilvum_PYR-GCK LLDERIEAIKVLWTQEPAEYHGRFVDFDASYQRPKPVRKPHPPILIGGDS
Mvan_5819|M.vanbaalenii_PYR-1 LLDERIEAIKALWTAEPAEYHGKYVDFDASYLRPKPVQKPHPPILIGGDS
TH_2750|M.thermoresistible__bu LLDERIEAVKALWTDEPAEYHGRFVDFGPSYLRPKPVQKPHPPIVVGGNS
MMAR_3559|M.marinum_M LLDERIEAIKALWTDEPAEYHGKYVDFPPSYSRPKPVRQPHPPIYIGGNS
MUL_2792|M.ulcerans_Agy99 LLDERIEAIKALWTDEPAEYHGKYVDFPPSYSRPKPVRQPHPPIYIGGNS
Mb3106c|M.bovis_AF2122/97 VIDERLRAIIEIWTQEQAEFHGTYVDFDPIYCWPKPVTKPYPPLYVGGGP
Rv3079c|M.tuberculosis_H37Rv VIDERLRAIIEIWTQEQAEFHGTYVDFDPIYCWPKPVTKPYPPLYVGGGP
MAV_1077|M.avium_104 MLREYLEAMRALWTQEEAAYDGEFVKFGPSWAWPKPVQPHIPVLVGAAGN
MLBr_00269|M.leprae_Br4923 RLRESVRLMRELWRGDRVDFDGDYYQLKGASIYDVPEGG--VPIYIAAGG
: * : : :* : . : * : * : ..
MAB_4908c|M.abscessus_ATCC_199 RAAQARALRVGDGWLPYAG---TSTPDDVRRACGWFAEQGRAD---LPVT
MSMEG_6703|M.smegmatis_MC2_155 DAALARTLRFGDGWLPFAE---LSTPADVARARKWFAEQGRND---LGVT
Mflv_1017|M.gilvum_PYR-GCK DATVRRIIRHGAGWIANP-----LPIDHLSRRIEQIREGAGGD---VPLC
Mvan_5819|M.vanbaalenii_PYR-1 DAAVKRIIRHEAGWISNP-----LPVDTLRRRVDQIRAGARHE---VPLS
TH_2750|M.thermoresistible__bu DATVRRIIRHGAGWMSNP-----HPPEMLKRRVQQIRDGAGHD---VLLT
MMAR_3559|M.marinum_M DATVKRVVRHDAGWISNP-----LPVRQLTQRVDQLRGAAGHD---VPLA
MUL_2792|M.ulcerans_Agy99 DATVKRVVRHDAGWISNP-----LPVHQLTQRVDQLRSAAGHD---VPLA
Mb3106c|M.bovis_AF2122/97 -ANFPRIARLNAGWIAIS-----PSPQRLSGPLQRLRAMAGGD---VPVT
Rv3079c|M.tuberculosis_H37Rv -ANFPRIARLNAGWIAIS-----PSPQRLSGPLQRLRAMAGGD---VPVT
MAV_1077|M.avium_104 EKNFKWIARSADGWITTPR---DFDIDEPVKLLQDTWAAAGRDG--APQI
MLBr_00269|M.leprae_Br4923 PEVAKYAGRAGEGFVCTSGKGEELYTEKLIPAVLEGAAVAGRDADDIDKM
* *:: . . :
MAB_4908c|M.abscessus_ATCC_199 VSGV-PGDHQLAGEYLGAGAE--------------------RVLFDVHPL
MSMEG_6703|M.smegmatis_MC2_155 VSGV-PGDASVASAYLDAGAG--------------------RVLFNLEPG
Mflv_1017|M.gilvum_PYR-GCK QFGT-PVKPEYWQAAEELGFG--------------------QLNLLLPTK
Mvan_5819|M.vanbaalenii_PYR-1 MFGT-PVKPEYWQAAEDLGFG--------------------QLNLLLPTK
TH_2750|M.thermoresistible__bu TFGT-PVDPGYWEVLEELGYQ--------------------QANLLLPTR
MMAR_3559|M.marinum_M MFGT-PNKPDYWRAAQDLGFG--------------------QLALLLPT-
MUL_2792|M.ulcerans_Agy99 MFGT-RNKPDYWRAAQDLGFGP-----------------AGAAAAHLPAG
Mb3106c|M.bovis_AF2122/97 VCQWGEAAAKDLEGYRHLGVE--------------------RVLLELPTE
Rv3079c|M.tuberculosis_H37Rv VCQWGEAAAKDLEGYRHLGVE--------------------RVLLELPTE
MAV_1077|M.avium_104 VALDFKPDAEKLARWAELGVT--------------------EVLFGLPDK
MLBr_00269|M.leprae_Br4923 IEIKMSYDPDPEQALSNIRFWAPLSLAAEQKHSIDDPIEMEKVADALPIE
:
MAB_4908c|M.abscessus_ATCC_199 -----------NEA-----------------------DTLRALDRLTEVV
MSMEG_6703|M.smegmatis_MC2_155 -----------PED-----------------------DVLRALDTVADTV
Mflv_1017|M.gilvum_PYR-GCK -----------SRD-----------------------VSLRLLDDYAEQV
Mvan_5819|M.vanbaalenii_PYR-1 -----------PRD-----------------------ESLRLLDGYAEQV
TH_2750|M.thermoresistible__bu -----------PRD-----------------------ESLRMLDQFAERV
MMAR_3559|M.marinum_M ----------CPRD-----------------------ESLRLLDEYAALV
MUL_2792|M.ulcerans_Agy99 GVPAAARRIRCPGGSIPPV---------------EVPGGPSFLREPLARV
Mb3106c|M.bovis_AF2122/97 -----------PRD-----------------------PTLRYLDKLQAEL
Rv3079c|M.tuberculosis_H37Rv -----------PRD-----------------------PTLRYLDKLQAEL
MAV_1077|M.avium_104 -----------SED-----------------------EVAAYVERLAGKL
MLBr_00269|M.leprae_Br4923 QVAKRWIVVSDPDEAVARVGQYVTWGLNHLVFHAPGHNQRRFLELFEKDL
: :
MAB_4908c|M.abscessus_ATCC_199 ARLT-----------------------------
MSMEG_6703|M.smegmatis_MC2_155 RSI------------------------------
Mflv_1017|M.gilvum_PYR-GCK ARYIG----------------------------
Mvan_5819|M.vanbaalenii_PYR-1 ARY------------------------------
TH_2750|M.thermoresistible__bu AGYRG----------------------------
MMAR_3559|M.marinum_M AQYHR----------------------------
MUL_2792|M.ulcerans_Agy99 SCASRRKIAPRFAAQCDFASARGGRLRVGSRPR
Mb3106c|M.bovis_AF2122/97 ARLA-----------------------------
Rv3079c|M.tuberculosis_H37Rv ARLA-----------------------------
MAV_1077|M.avium_104 AALV-----------------------------
MLBr_00269|M.leprae_Br4923 APRLRRLG-------------------------