For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
THPDRANVNPGSPPLRETLSQLRLRELLLEVQDRIEQIVEGRDRLDGLIDAILAITSGLKLDATLRAIVH TAAELVDARYGALGVRGYDHRLVEFVYEGIDEETRHLIGSLPEGRGVLGALIEEPKPIRLDDISRHPASV GFPLHHPPMRTFLGVPVRIRDEVFGNLYLTEKADGQPFSDDDEVLVQALAAAAGIAVDNARLFEESRTRE AWIEATRDIGTQMLAGADPAMVFRLIAEEALTLMAGAATLVAVPLDDEAPACEVDDLVIVEVAGEISPAV KQMTVAVSGTSIGGVFHDRTPRRFDRLDLAVDGPVEPGPALVLPLRAADTVAGVLVALRSADEQPFSDKQ LDMMAAFADQAALAWRLATAQRQMREVEILTDRDRIARDLHDHVIQRLFAVGLTLQGAAPRARVPAVRES IYSSIDDLQEIIQEIRSAIFDLHAGPSRATGLRHRLDKVIDQLAIPALHTTVQYTGPLSVVDTVLANHAE AVLREAVSNAVRHANATSLAINVSVEDDVRVEVVDDGVGISGDITESGLRNLRQRADDAGGEFTVENMPT GGTLLRWSAPLR*
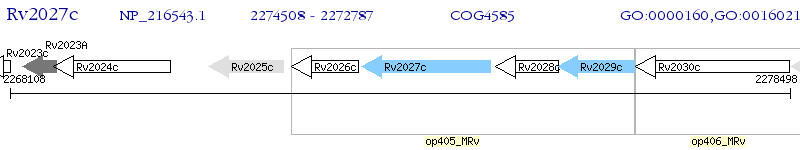
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2027c | - | - | 100% (573) | Probable histidine kinase response regulator |
| M. tuberculosis H37Rv | Rv3132c | devS | 0.0 | 62.50% (560) | two component sensor histidine kinase DEVS |
| M. tuberculosis H37Rv | Rv0845 | - | 8e-11 | 28.57% (210) | two component sensor kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2052c | - | 0.0 | 99.83% (573) | histidine kinase response regulator |
| M. gilvum PYR-GCK | Mflv_3844 | - | 0.0 | 63.64% (572) | GAF sensor signal transduction histidine kinase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3890c | - | 1e-156 | 53.26% (567) | histidine kinase response regulator |
| M. marinum M | MMAR_1517 | devS | 0.0 | 61.25% (578) | two-component sensor histidine kinase DevS |
| M. avium 104 | MAV_4108 | - | 1e-121 | 43.85% (545) | GAF family protein |
| M. smegmatis MC2 155 | MSMEG_5241 | - | 0.0 | 68.28% (558) | GAF family protein |
| M. thermoresistible (build 8) | TH_0388 | - | 1e-76 | 58.16% (239) | TWO COMPONENT SENSOR HISTIDINE KINASE DEVS |
| M. ulcerans Agy99 | MUL_2422 | devS | 0.0 | 61.07% (578) | two component sensor histidine kinase DevS |
| M. vanbaalenii PYR-1 | Mvan_1395 | - | 0.0 | 67.80% (559) | GAF sensor signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2027c|M.tuberculosis_H37Rv ---------MTHPDRANVNPGSPPLRETLSQLRLRELLLEVQDRIEQIVE
Mb2052c|M.bovis_AF2122/97 ---------MTHPDRANVNPGSPPVRETLSQLRLRELLLEVQDRIEQIVE
Mflv_3844|M.gilvum_PYR-GCK ----MHDGAVADADDHRARDGARPVRTTLSQLRLRELLTEVQDRVEQIIR
Mvan_1395|M.vanbaalenii_PYR-1 ---------------------MRPLRDTLSQLRLRELLNEVQDRVEQIVE
MSMEG_5241|M.smegmatis_MC2_155 ------MGGVNGHHPRRDDDQRTPLRDTLSQLRLRELLSEVQDRVEQIVE
MMAR_1517|M.marinum_M MTDEPHPQDAGPALGVPSGDGLRPLRLTLSQLRLRELLVEVQERVEQIVE
MUL_2422|M.ulcerans_Agy99 MTDEPHPQDAGPSLGVPSGDGLRPLRLTLSQLRLRELLVEVQERVEQIVE
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 -------------MVEDVGAQPNRVTAELAHLQLRELLAEVQGRIEQIVD
MAV_4108|M.avium_104 ---------------------------------------MTGNRFDELAA
Rv2027c|M.tuberculosis_H37Rv G-RDRLDGLIDAILAITSGLKLDATLRAIVHTAAELVDARYGALGVRGYD
Mb2052c|M.bovis_AF2122/97 G-RDRLDGLIDAILAITSGLKLDATLRAIVHTAAELVDARYGALGVRGYD
Mflv_3844|M.gilvum_PYR-GCK G-RDRLDGLVEAMLAVTSGLELDETLRTIVHTAIELVDAQYGALGVRGHD
Mvan_1395|M.vanbaalenii_PYR-1 G-RDRLDGLLDAMLVVTSGLELDETLRTIVRTAIDLVDAQYGALGVRGHD
MSMEG_5241|M.smegmatis_MC2_155 G-RDRLDGLVEAMLVVTSGLELDVTLRTIVHTAIELVDARYGALGVRGED
MMAR_1517|M.marinum_M G-RDRLDGLVEAMLVVTSGLELNATLRTIVHSATNLVDARYCALEVHDRD
MUL_2422|M.ulcerans_Agy99 G-RDRLDGLVEAMLVVTSGLELDATLRTIVHSATNLVDARYCALEVHDRD
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 GTRGRMDALLDAVMAVSSGLELDATLREIVCAASELVSARYGALGVVGSD
MAV_4108|M.avium_104 A-RDQTEKLLRVIAEIGAGLDLDATLHRIISAARELTSAPYGALAVRDWH
Rv2027c|M.tuberculosis_H37Rv HRLVEFVYEGIDEETRHLIGSLPEGRGVLGALIEEPKPIRLDDISRHPAS
Mb2052c|M.bovis_AF2122/97 HRLVEFVYEGIDEETRHLIGSLPEGRGVLGALIEEPKPIRLDDISRHPAS
Mflv_3844|M.gilvum_PYR-GCK QELVEFIYEGIDEPMRERIGHLPEGRGVLGVLINDPKPIRLEDISRHADS
Mvan_1395|M.vanbaalenii_PYR-1 HELVEFIYQGIDEQSKELIGHLPEGRGVLGVLIDDPKPIRLDRISQHPAS
MSMEG_5241|M.smegmatis_MC2_155 HHLIEFIYEGITPEVREEIGPLPEGRGVLGVLIDDPKPIRLDDIRLHPAS
MMAR_1517|M.marinum_M KRVLQFVYEGIDEDTVARIGHLPEGLGVIGLLIDEPKPLRLEDISQHPAS
MUL_2422|M.ulcerans_Agy99 KRVLQFVYEGIDEDTVARIGHLPEGLGIIGLLIDEPKPLRLEDISQHPAS
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 EKLAEFVYEGIDEATRDLIGPLPTGHGVLGVVIDEAKPLRLRDIAAHPAS
MAV_4108|M.avium_104 ANLISFVHEGMDAETVRRIGHHPVGKGLLSLSLLDTPALRMDDLTAHPAA
Rv2027c|M.tuberculosis_H37Rv VGFPLHHPPMRTFLGVPVRIRDEVFGNLYLTEKADGQPFSDDDEVLVQAL
Mb2052c|M.bovis_AF2122/97 VGFPLHHPPMRTFLGVPVRIRDEVFGNLYLTEKADGQPFSDDDEVLVQAL
Mflv_3844|M.gilvum_PYR-GCK VGFPPNHPPMRTFLGVPVRIRDEVFGNLYLTEKSDGQPFSEDDEVLVQAL
Mvan_1395|M.vanbaalenii_PYR-1 VGFPPNHPPMRTFLGVPVRIRDEVFGNLYLTEKADGQPFSEDDEVLVQAL
MSMEG_5241|M.smegmatis_MC2_155 VGFPPNHPPMRTFLGVPVRTRDKVFGNLYLTEKTNGQPFSEDDEVLVQAL
MMAR_1517|M.marinum_M IGFPSHHPPMRTFLGVPVRVRDVSLGTLYLTDKTNGQPFSDDDELLVQAL
MUL_2422|M.ulcerans_Agy99 IGFPSHHPPMRTFLGVPVRVRDVSLGTLYLTDKTNGQPFSDDDELLVQAL
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 VGFPANHPPMRTFLGVPVQVRGEVFGRLYLTEKLDGQEFTEDDEVVVRAL
MAV_4108|M.avium_104 AGFPEHHPAMRAFLAVPITIRGAVFGNLYLTHVDEARVFSDADEMAARAL
Rv2027c|M.tuberculosis_H37Rv AAAAGIAVDNARLFEESRTREAWIEATRDIGTQMLAGADPAM-VFRLIAE
Mb2052c|M.bovis_AF2122/97 AAAAGIAVDNARLFEESRTREAWIEATRDIGTQMLAGADPAM-VFRLIAE
Mflv_3844|M.gilvum_PYR-GCK AAAAGIAIDNARLYAQSQARQSWIEATRDIGTELLSGIDPAG-VFRRVAD
Mvan_1395|M.vanbaalenii_PYR-1 AAAAGIAIENARLYQQSKTRRSWIEATRDIGTEMLSGADPSK-VFRLVAD
MSMEG_5241|M.smegmatis_MC2_155 AAAAGIAVDNARLYEESQARQAWIAATRDIGTQLLSGTDPAT-VFRLVAA
MMAR_1517|M.marinum_M AAAAGIAIANARLYEQAKARQAWIGATRDIATELLSGTEPAS-VFRLVAE
MUL_2422|M.ulcerans_Agy99 AAAAGIAIANARLYEQAKARQAWIGATRDIATELLSGTEPAS-VFRLVAE
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 AGAAGIAIENARLYQHARRREQWQRATADITAELLGGVDPGK-ALHLVAS
MAV_4108|M.avium_104 AFAAAVAIDNAQVFERERMSVKWIEASREITTALLSRTEPHRRPMQLIAE
Rv2027c|M.tuberculosis_H37Rv EALTLMAGAATLVAVPLDDEAPAC-EVDDLVIVEVAGEISPAVKQMTVAV
Mb2052c|M.bovis_AF2122/97 EALTLMAGAATLVAVPLDDEAPAC-EVDDLVIVEVAGEISPAVKQMTVAV
Mflv_3844|M.gilvum_PYR-GCK RAQKLSGAAATLVAVPDDPDLPVG-EVAELTVAASAGET-IAAAHDPIPV
Mvan_1395|M.vanbaalenii_PYR-1 QSRLLSGAQSTLVAVRSDLDELDG-RVDELVVVATAGDG-PGVSLSAIPT
MSMEG_5241|M.smegmatis_MC2_155 EALTLTGADGTLVAVPADPDASA---AEELVIVEVAGAVPAEVEASAIPV
MMAR_1517|M.marinum_M EALKLTSADAALVAVPVDEDVPAS-EVAQLLVIETVGNAVAAAEGCTIAV
MUL_2422|M.ulcerans_Agy99 EALKLTSADAALVAIPVDEDVPAS-EVAQLLVIETVGNAVAAAEGCTIAV
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 SAALLAEADFAFIALPAGD---EE-DTAELVVAVCEGKGAEVLNDKLIPI
MAV_4108|M.avium_104 RACALTDAEQAIVLVPADPEQPGNPEVDTLVVSAAVGLPAADVLGQRVPV
Rv2027c|M.tuberculosis_H37Rv SGTSIGGVFHDRTPRRFDRLDLAVDGPV--EPGPALVLPLRAADTVAGVL
Mb2052c|M.bovis_AF2122/97 SGTSIGGVFHDRTPRRFDRLDLAVDGPV--EPGPALVLPLRAADTVAGVL
Mflv_3844|M.gilvum_PYR-GCK AGTAIGDAFVHRTPGMTDRADIG--IGA--TVGPALLLPLRAPDAVPGVL
Mvan_1395|M.vanbaalenii_PYR-1 HGSAIGEVFSTKTPVRFDTLELEP-RHL--APGPALVLPLRATDTVAGVL
MSMEG_5241|M.smegmatis_MC2_155 QDNAIGQAFRDRAPRRLDVLDGPG------LGGPALVLPLRATDTVAGVL
MMAR_1517|M.marinum_M AGTALAEVLLDSAPRQVDKIAVEGVDELG-DTGPALVLPLRATDAVAGVV
MUL_2422|M.ulcerans_Agy99 AGTALAEVLLDSAPRQVDKIAVEDVDELG-DMGPALVLPLRATDAVAGVV
TH_0388|M.thermoresistible__bu ------------------------------------------------VL
MAB_3890c|M.abscessus_ATCC_199 DGSTSGSTFIDHEPRNVGRLAVNLAEGTDLAFGPALVLPLGGGESTAGVL
MAV_4108|M.avium_104 RGSTSGAVFKSGEPLITDEFRYPIQAFTDAGRRPAIVMPLRARDEVVGVI
*:
Rv2027c|M.tuberculosis_H37Rv VALRSADEQPFSDKQLDMMAAFADQAALAWRLATAQRQMREVEILTDRDR
Mb2052c|M.bovis_AF2122/97 VALRSADEQPFSDKQLDMMAAFADQAALAWRLATAQRQMREVEILTDRDR
Mflv_3844|M.gilvum_PYR-GCK IVLRSVGAQPFRSDELDMLAAFADQAAQAWQLASTQHRLRELGILTDRDR
Mvan_1395|M.vanbaalenii_PYR-1 VALRPEGAPPFNAEELDMMAAFVDQAALAWQLATTQRQLRELDVLTDRDR
MSMEG_5241|M.smegmatis_MC2_155 VAVQGSGARPFTAEQLEMMTGFADQAAVAWQLASSQRRMSELDILADRDR
MMAR_1517|M.marinum_M VLLHPSSPESVTEEQLEMMAAFADQAALALQLATSQRRMRELDVMTERDR
MUL_2422|M.ulcerans_Agy99 VLLHPSSPESVTEEQLEMMAAFADQAALALQLATSQRRMRELDVVTERDR
TH_0388|M.thermoresistible__bu VVCRAEEGRPFTDEQLEILVTFADQAALAWQLAATQRRMRELDVLSDRDR
MAB_3890c|M.abscessus_ATCC_199 ILLRTVGAIAFDEEQLDMAASFAGQAALALEQAEVRLAKHELDVLADRDR
MAV_4108|M.avium_104 AIARGTEQPPFDESYLDLVRDFATHAAIALVLAAAREDARQLAVLAERER
: .. . *:: *. :** * * : :: ::::*:*
Rv2027c|M.tuberculosis_H37Rv IARDLHDHVIQRLFAVGLTLQGAAPRARVPAVRESIYSSIDDLQEIIQEI
Mb2052c|M.bovis_AF2122/97 IARDLHDHVIQRLFAVGLTLQGAAPRARVPAVRESIYSSIDDLQEIIQEI
Mflv_3844|M.gilvum_PYR-GCK IARDLHDHVIQRLFAVGLSLQGSIARARSEEVQQRLSANVDDLQSVIQEI
Mvan_1395|M.vanbaalenii_PYR-1 IARDLHDHVIQRLFAVGLALQGTIPRARAPEVRERLTDCVDDLQQVIQEI
MSMEG_5241|M.smegmatis_MC2_155 IARDLHDHVIQRLFAVGLSLQGTIPRARSAEVQQRIADCVDDLQAVISEI
MMAR_1517|M.marinum_M IARDLHDHVIQRLFAIGLALQGSVPRTREPKVQQRLSEVIDDLQGVIQEI
MUL_2422|M.ulcerans_Agy99 IARDLHDHVIQRLFAIGLALQGSVPRTREPKVQQRLSEVIDDLQGVIQEI
TH_0388|M.thermoresistible__bu IARDLHDHVIQRLFAVGLTLQGTIPRARSTEVQQRLSECIDDLQAVIGEI
MAB_3890c|M.abscessus_ATCC_199 IARDLHDHVIQRLFAVGLAMQSTHRRSVDPTVAARLADHIDQLHEVIQEI
MAV_4108|M.avium_104 IAHDLHDHVIQRLFAAGMDLQGTLARARSPEVADRLNRTLDDLQTIIEEI
**:************ *: :*.: *: * : :*:*: :* **
Rv2027c|M.tuberculosis_H37Rv RSAIFDLHAGPSRATGLRHRLDKVIDQLAIP-ALHTTVQYTGPLSVVDTV
Mb2052c|M.bovis_AF2122/97 RSAIFDLHAGPSRATGLRHRLDKVIDQLAIP-ALHTTVQYTGPLSVVDTV
Mflv_3844|M.gilvum_PYR-GCK RTAIFDLHGGSSAVTRLRQRLDAAVAAFPES-GVRTSMQCAGPLSVVDAG
Mvan_1395|M.vanbaalenii_PYR-1 RTAIFDLHGGHGGSNRLRQRLDEAIAQFTAS-DVHTTVQFSGPLSVVDPA
MSMEG_5241|M.smegmatis_MC2_155 RTAIFDLHHTAPGGTRLRQRLDEAIAQFSGA-GLHTTSQFVGPLSVVDAE
MMAR_1517|M.marinum_M RTTIFDLHGSSQGVTRLRQRIDAAVAAFGSS-GLRTTVQFIGPLSVVDNV
MUL_2422|M.ulcerans_Agy99 RTTIFDLHGSSQGVTRLRQRIDAAVAAFGSS-GLRTTVQFIGPLSVVDNV
TH_0388|M.thermoresistible__bu RTTIFDLHGGPATGSQIRQRIGEAANQFSDA-GVRTRVQFAGPLSVISAV
MAB_3890c|M.abscessus_ATCC_199 RTAIFDLHTNDES---LRATLRSTINELTAETGLRVSVRMSGPLDVVPAS
MAV_4108|M.avium_104 RTTIFALRSPAAVAGDFRHRIQRVIAELTENRDLVTTVRMDGPMTAVGAE
*::** *: :* : . : : . : **: .:
Rv2027c|M.tuberculosis_H37Rv LANHAEAVLREAVSNAVRHANATSLAINVSVEDDVRVEVVDDGVGIS-GD
Mb2052c|M.bovis_AF2122/97 LANHAEAVLREAVSNAVRHANATSLAINVSVEDDVRVEVVDDGVGIS-GD
Mflv_3844|M.gilvum_PYR-GCK LADHAEAVVREAVSNAVRHAEASTVAVTVTVEDDLAITVTDDGRGIA-AD
Mvan_1395|M.vanbaalenii_PYR-1 LSDHAEAVVREAVSNAIRHSGATTLDVSVTAADELIIEVADDGCGMP-AD
MSMEG_5241|M.smegmatis_MC2_155 LADHAEAVLREAVSNVVRHSGANELTVRVKVEDDLSIEVTDDGEGIS-GP
MMAR_1517|M.marinum_M LADHAEAVVREAVSNAVRHAQATTLTVRVKVDDDLCIEVSDNGRGMP-KT
MUL_2422|M.ulcerans_Agy99 LADHAEAVVREAVSNAVRHAQATTLTVRVKVDDDLCIEVSDNGRGMP-KT
TH_0388|M.thermoresistible__bu LADHAEAVVREAVSNAVRHGGATDLSVDVSVDDDLTIEVTDNGRGFPEGE
MAB_3890c|M.abscessus_ATCC_199 TSGDAEAVVREAVSNVVRHARARELSVTVSVDDELVIEVVDDGIGIP-DV
MAV_4108|M.avium_104 LAEHAEAVTAEAVSNAVRHSGASRLTVQIGVADMFTLDVIDNGRGIAAGN
: .**** *****.:**. * : : : . * . : * *:* *:.
Rv2027c|M.tuberculosis_H37Rv ITESGLRNLRQRADDAGGEFTVEN-MPTGGTLLRWSAPLR--
Mb2052c|M.bovis_AF2122/97 ITESGLRNLRQRADDAGGEFTVEN-MPTGGTLLRWSAPLR--
Mflv_3844|M.gilvum_PYR-GCK VTGSGLANLARRAQDVGGSFSVRR-GAQVGTTLTWRAPLP--
Mvan_1395|M.vanbaalenii_PYR-1 VTESGLTNLRQRAGDVGGTFTVES-GDAGGTRLRWCAPLR--
MSMEG_5241|M.smegmatis_MC2_155 VTESGLSNLRQRADQCGGELRIID-RPGGGTILRWTAPLPD-
MMAR_1517|M.marinum_M YTSSGLTNLRRRAQEAGGQFTIESPAAGGGTLLRWSAPLIQ-
MUL_2422|M.ulcerans_Agy99 YTSSGLTNLRRRAQEAGGQFTIESPAAGGGTLLRWSAPLIQ-
TH_0388|M.thermoresistible__bu VTESGLANLRHRARHAGGTLEVHN-RPDGGARLHWSVPLT--
MAB_3890c|M.abscessus_ATCC_199 VARSGLSGLANRAAQSGGTFSVSA-MDTGGTRLVWSAPLP--
MAV_4108|M.avium_104 TRRSGLANMTRRAEQLGGSCEISS-PPGGGTRVHWTAPLLDH
*** .: .** . ** : *: : * .**