For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HDGAVADADDHRARDGARPVRTTLSQLRLRELLTEVQDRVEQIIRGRDRLDGLVEAMLAVTSGLELDETL RTIVHTAIELVDAQYGALGVRGHDQELVEFIYEGIDEPMRERIGHLPEGRGVLGVLINDPKPIRLEDISR HADSVGFPPNHPPMRTFLGVPVRIRDEVFGNLYLTEKSDGQPFSEDDEVLVQALAAAAGIAIDNARLYAQ SQARQSWIEATRDIGTELLSGIDPAGVFRRVADRAQKLSGAAATLVAVPDDPDLPVGEVAELTVAASAGE TIAAAHDPIPVAGTAIGDAFVHRTPGMTDRADIGIGATVGPALLLPLRAPDAVPGVLIVLRSVGAQPFRS DELDMLAAFADQAAQAWQLASTQHRLRELGILTDRDRIARDLHDHVIQRLFAVGLSLQGSIARARSEEVQ QRLSANVDDLQSVIQEIRTAIFDLHGGSSAVTRLRQRLDAAVAAFPESGVRTSMQCAGPLSVVDAGLADH AEAVVREAVSNAVRHAEASTVAVTVTVEDDLAITVTDDGRGIAADVTGSGLANLARRAQDVGGSFSVRRG AQVGTTLTWRAPLP*
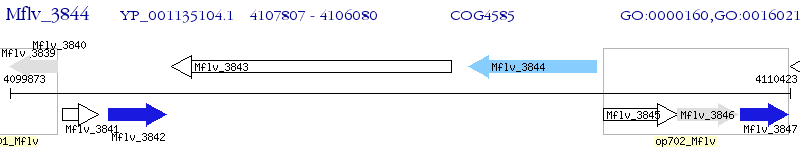
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3844 | - | - | 100% (575) | GAF sensor signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_4521 | - | 3e-44 | 35.42% (336) | putative GAF sensor protein |
| M. gilvum PYR-GCK | Mflv_5015 | - | 7e-17 | 29.95% (207) | GAF sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2052c | - | 0.0 | 63.81% (572) | histidine kinase response regulator |
| M. tuberculosis H37Rv | Rv2027c | - | 0.0 | 63.64% (572) | histidine kinase response regulator |
| M. leprae Br4923 | MLBr_00803 | - | 8e-05 | 22.79% (215) | putative two-component system sensor kinase |
| M. abscessus ATCC 19977 | MAB_3890c | - | 1e-151 | 53.17% (568) | histidine kinase response regulator |
| M. marinum M | MMAR_1517 | devS | 0.0 | 64.36% (564) | two-component sensor histidine kinase DevS |
| M. avium 104 | MAV_4108 | - | 1e-114 | 42.94% (545) | GAF family protein |
| M. smegmatis MC2 155 | MSMEG_5241 | - | 0.0 | 67.77% (574) | GAF family protein |
| M. thermoresistible (build 8) | TH_0388 | - | 6e-77 | 61.92% (239) | TWO COMPONENT SENSOR HISTIDINE KINASE DEVS |
| M. ulcerans Agy99 | MUL_2422 | devS | 0.0 | 64.18% (564) | two component sensor histidine kinase DevS |
| M. vanbaalenii PYR-1 | Mvan_1395 | - | 0.0 | 70.92% (557) | GAF sensor signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3844|M.gilvum_PYR-GCK ----MHDGAVADADDHRARDGARPVRTTLSQLRLRELLTEVQDRVEQIIR
Mvan_1395|M.vanbaalenii_PYR-1 ---------------------MRPLRDTLSQLRLRELLNEVQDRVEQIVE
Mb2052c|M.bovis_AF2122/97 ---------MTHPDRANVNPGSPPVRETLSQLRLRELLLEVQDRIEQIVE
Rv2027c|M.tuberculosis_H37Rv ---------MTHPDRANVNPGSPPLRETLSQLRLRELLLEVQDRIEQIVE
MSMEG_5241|M.smegmatis_MC2_155 ----MGGVNGHHPRRD--DDQRTPLRDTLSQLRLRELLSEVQDRVEQIVE
MMAR_1517|M.marinum_M MTDEPHPQDAGPALGVPSGDGLRPLRLTLSQLRLRELLVEVQERVEQIVE
MUL_2422|M.ulcerans_Agy99 MTDEPHPQDAGPSLGVPSGDGLRPLRLTLSQLRLRELLVEVQERVEQIVE
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 -------------MVEDVGAQPNRVTAELAHLQLRELLAEVQGRIEQIVD
MAV_4108|M.avium_104 ---------------------------------------MTGNRFDELAA
MLBr_00803|M.leprae_Br4923 --------------------------------------------------
Mflv_3844|M.gilvum_PYR-GCK G-RDRLDGLVEAMLAVTSGLELDETLRTIVHTAIELVDAQYGALGVRGHD
Mvan_1395|M.vanbaalenii_PYR-1 G-RDRLDGLLDAMLVVTSGLELDETLRTIVRTAIDLVDAQYGALGVRGHD
Mb2052c|M.bovis_AF2122/97 G-RDRLDGLIDAILAITSGLKLDATLRAIVHTAAELVDARYGALGVRGYD
Rv2027c|M.tuberculosis_H37Rv G-RDRLDGLIDAILAITSGLKLDATLRAIVHTAAELVDARYGALGVRGYD
MSMEG_5241|M.smegmatis_MC2_155 G-RDRLDGLVEAMLVVTSGLELDVTLRTIVHTAIELVDARYGALGVRGED
MMAR_1517|M.marinum_M G-RDRLDGLVEAMLVVTSGLELNATLRTIVHSATNLVDARYCALEVHDRD
MUL_2422|M.ulcerans_Agy99 G-RDRLDGLVEAMLVVTSGLELDATLRTIVHSATNLVDARYCALEVHDRD
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 GTRGRMDALLDAVMAVSSGLELDATLREIVCAASELVSARYGALGVVGSD
MAV_4108|M.avium_104 A-RDQTEKLLRVIAEIGAGLDLDATLHRIISAARELTSAPYGALAVRDWH
MLBr_00803|M.leprae_Br4923 --MSTLGDLLAEHTVLPG--NAVGHLHAVVGEWQLLADLSFADYLMWVSR
Mflv_3844|M.gilvum_PYR-GCK QELVEFIYEGIDEPMRERIGHLPEGRGVLGVLINDPKPIRLEDISRHADS
Mvan_1395|M.vanbaalenii_PYR-1 HELVEFIYQGIDEQSKELIGHLPEGRGVLGVLIDDPKPIRLDRISQHPAS
Mb2052c|M.bovis_AF2122/97 HRLVEFVYEGIDEETRHLIGSLPEGRGVLGALIEEPKPIRLDDISRHPAS
Rv2027c|M.tuberculosis_H37Rv HRLVEFVYEGIDEETRHLIGSLPEGRGVLGALIEEPKPIRLDDISRHPAS
MSMEG_5241|M.smegmatis_MC2_155 HHLIEFIYEGITPEVREEIGPLPEGRGVLGVLIDDPKPIRLDDIRLHPAS
MMAR_1517|M.marinum_M KRVLQFVYEGIDEDTVARIGHLPEGLGVIGLLIDEPKPLRLEDISQHPAS
MUL_2422|M.ulcerans_Agy99 KRVLQFVYEGIDEDTVARIGHLPEGLGIIGLLIDEPKPLRLEDISQHPAS
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 EKLAEFVYEGIDEATRDLIGPLPTGHGVLGVVIDEAKPLRLRDIAAHPAS
MAV_4108|M.avium_104 ANLISFVHEGMDAETVRRIGHHPVGKGLLSLSLLDTPALRMDDLTAHPAA
MLBr_00803|M.leprae_Br4923 DDGVLVCVAQCRPHT-------------------APTVLQTDAVGSVASA
Mflv_3844|M.gilvum_PYR-GCK VGFPPNHPPMRTFLGVPVRIRDEVFGNLYLTEKSDGQPFSEDDEVLVQAL
Mvan_1395|M.vanbaalenii_PYR-1 VGFPPNHPPMRTFLGVPVRIRDEVFGNLYLTEKADGQPFSEDDEVLVQAL
Mb2052c|M.bovis_AF2122/97 VGFPLHHPPMRTFLGVPVRIRDEVFGNLYLTEKADGQPFSDDDEVLVQAL
Rv2027c|M.tuberculosis_H37Rv VGFPLHHPPMRTFLGVPVRIRDEVFGNLYLTEKADGQPFSDDDEVLVQAL
MSMEG_5241|M.smegmatis_MC2_155 VGFPPNHPPMRTFLGVPVRTRDKVFGNLYLTEKTNGQPFSEDDEVLVQAL
MMAR_1517|M.marinum_M IGFPSHHPPMRTFLGVPVRVRDVSLGTLYLTDKTNGQPFSDDDELLVQAL
MUL_2422|M.ulcerans_Agy99 IGFPSHHPPMRTFLGVPVRVRDVSLGTLYLTDKTNGQPFSDDDELLVQAL
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 VGFPANHPPMRTFLGVPVQVRGEVFGRLYLTEKLDGQEFTEDDEVVVRAL
MAV_4108|M.avium_104 AGFPEHHPAMRAFLAVPITIRGAVFGNLYLTHVDEARVFSDADEMAARAL
MLBr_00803|M.leprae_Br4923 DMLPLVTEMFTSGAVQHEDAAGQQYSWLPFGRNVAASPVRYGDQVVAVLT
Mflv_3844|M.gilvum_PYR-GCK AAAAGIAIDNARLYAQSQARQSWIEATRDIGTELLSGIDPAG-VFRRVAD
Mvan_1395|M.vanbaalenii_PYR-1 AAAAGIAIENARLYQQSKTRRSWIEATRDIGTEMLSGADPSK-VFRLVAD
Mb2052c|M.bovis_AF2122/97 AAAAGIAVDNARLFEESRTREAWIEATRDIGTQMLAGADPAM-VFRLIAE
Rv2027c|M.tuberculosis_H37Rv AAAAGIAVDNARLFEESRTREAWIEATRDIGTQMLAGADPAM-VFRLIAE
MSMEG_5241|M.smegmatis_MC2_155 AAAAGIAVDNARLYEESQARQAWIAATRDIGTQLLSGTDPAT-VFRLVAA
MMAR_1517|M.marinum_M AAAAGIAIANARLYEQAKARQAWIGATRDIATELLSGTEPAS-VFRLVAE
MUL_2422|M.ulcerans_Agy99 AAAAGIAIANARLYEQAKARQAWIGATRDIATELLSGTEPAS-VFRLVAE
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 AGAAGIAIENARLYQHARRREQWQRATADITAELLGGVDPGK-ALHLVAS
MAV_4108|M.avium_104 AFAAAVAIDNAQVFERERMSVKWIEASREITTALLSRTEPHRRPMQLIAE
MLBr_00803|M.leprae_Br4923 QHQVELAAR----RRPGNLEIAYLACATDLLHMLAEGT------FPDVGD
Mflv_3844|M.gilvum_PYR-GCK RAQKLSGAAATLVAVPDDPDLPVG-EVAELTVAASAGET-IAAAHDPIPV
Mvan_1395|M.vanbaalenii_PYR-1 QSRLLSGAQSTLVAVRSDLDELDG-RVDELVVVATAGDG-PGVSLSAIPT
Mb2052c|M.bovis_AF2122/97 EALTLMAGAATLVAVPLDDEAPAC-EVDDLVIVEVAGEISPAVKQMTVAV
Rv2027c|M.tuberculosis_H37Rv EALTLMAGAATLVAVPLDDEAPAC-EVDDLVIVEVAGEISPAVKQMTVAV
MSMEG_5241|M.smegmatis_MC2_155 EALTLTGADGTLVAVPADPDASAA-E--ELVIVEVAGAVPAEVEASAIPV
MMAR_1517|M.marinum_M EALKLTSADAALVAVPVDEDVPAS-EVAQLLVIETVGNAVAAAEGCTIAV
MUL_2422|M.ulcerans_Agy99 EALKLTSADAALVAIPVDEDVPAS-EVAQLLVIETVGNAVAAAEGCTIAV
TH_0388|M.thermoresistible__bu --------------------------------------------------
MAB_3890c|M.abscessus_ATCC_199 SAALLAEADFAFIALPAGDEE----DTAELVVAVCEGKGAEVLNDKLIPI
MAV_4108|M.avium_104 RACALTDAEQAIVLVPADPEQPGNPEVDTLVVSAAVGLPAADVLGQRVPV
MLBr_00803|M.leprae_Br4923 VAVSHSTPRAGDGFIRLDRNG-------VVAFASPNALSAFHRMGLTAEL
Mflv_3844|M.gilvum_PYR-GCK AGTAIGDAFVHRTPGMTDRADI-G-IGATV--GPALLLPLRAPDAVPGVL
Mvan_1395|M.vanbaalenii_PYR-1 HGSAIGEVFSTKTPVRFDTLEL-EPRHLAP--GPALVLPLRATDTVAGVL
Mb2052c|M.bovis_AF2122/97 SGTSIGGVFHDRTPRRFDRLDL-AVDGPVEP-GPALVLPLRAADTVAGVL
Rv2027c|M.tuberculosis_H37Rv SGTSIGGVFHDRTPRRFDRLDL-AVDGPVEP-GPALVLPLRAADTVAGVL
MSMEG_5241|M.smegmatis_MC2_155 QDNAIGQAFRDRAPRRLD-----VLDGPGLG-GPALVLPLRATDTVAGVL
MMAR_1517|M.marinum_M AGTALAEVLLDSAPRQVDKIAVEGVDELGDT-GPALVLPLRATDAVAGVV
MUL_2422|M.ulcerans_Agy99 AGTALAEVLLDSAPRQVDKIAVEDVDELGDM-GPALVLPLRATDAVAGVV
TH_0388|M.thermoresistible__bu ------------------------------------------------VL
MAB_3890c|M.abscessus_ATCC_199 DGSTSGSTFIDHEPRNVGRLAVNLAEGTDLAFGPALVLPLGGGESTAGVL
MAV_4108|M.avium_104 RGSTSGAVFKSGEPLITDEFRYPIQAFTDAGRRPAIVMPLRARDEVVGVI
MLBr_00803|M.leprae_Br4923 QGRNLVKVTRPLLSDPFEAQEVAKHVQDLLDGGPSMRVEVNAG----GAT
.
Mflv_3844|M.gilvum_PYR-GCK IVLRSVGAQPFRSDELDMLAAFADQAAQAWQLASTQHRLRELGILTDRDR
Mvan_1395|M.vanbaalenii_PYR-1 VALRPEGAPPFNAEELDMMAAFVDQAALAWQLATTQRQLRELDVLTDRDR
Mb2052c|M.bovis_AF2122/97 VALRSADEQPFSDKQLDMMAAFADQAALAWRLATAQRQMREVEILTDRDR
Rv2027c|M.tuberculosis_H37Rv VALRSADEQPFSDKQLDMMAAFADQAALAWRLATAQRQMREVEILTDRDR
MSMEG_5241|M.smegmatis_MC2_155 VAVQGSGARPFTAEQLEMMTGFADQAAVAWQLASSQRRMSELDILADRDR
MMAR_1517|M.marinum_M VLLHPSSPESVTEEQLEMMAAFADQAALALQLATSQRRMRELDVMTERDR
MUL_2422|M.ulcerans_Agy99 VLLHPSSPESVTEEQLEMMAAFADQAALALQLATSQRRMRELDVVTERDR
TH_0388|M.thermoresistible__bu VVCRAEEGRPFTDEQLEILVTFADQAALAWQLAATQRRMRELDVLSDRDR
MAB_3890c|M.abscessus_ATCC_199 ILLRTVGAIAFDEEQLDMAASFAGQAALALEQAEVRLAKHELDVLADRDR
MAV_4108|M.avium_104 AIARGTEQPPFDESYLDLVRDFATHAAIALVLAAAREDARQLAVLAERER
MLBr_00803|M.leprae_Br4923 VLLR----------TLPLVVHGSSVGAAVLVRDVTEVKRRDRALIS-KDA
: * : .* . . : ::: ::
Mflv_3844|M.gilvum_PYR-GCK IARDLHDHVIQRLFAVGLSLQGSIARARSEEVQQRLSANVDDLQSVIQEI
Mvan_1395|M.vanbaalenii_PYR-1 IARDLHDHVIQRLFAVGLALQGTIPRARAPEVRERLTDCVDDLQQVIQEI
Mb2052c|M.bovis_AF2122/97 IARDLHDHVIQRLFAVGLTLQGAAPRARVPAVRESIYSSIDDLQEIIQEI
Rv2027c|M.tuberculosis_H37Rv IARDLHDHVIQRLFAVGLTLQGAAPRARVPAVRESIYSSIDDLQEIIQEI
MSMEG_5241|M.smegmatis_MC2_155 IARDLHDHVIQRLFAVGLSLQGTIPRARSAEVQQRIADCVDDLQAVISEI
MMAR_1517|M.marinum_M IARDLHDHVIQRLFAIGLALQGSVPRTREPKVQQRLSEVIDDLQGVIQEI
MUL_2422|M.ulcerans_Agy99 IARDLHDHVIQRLFAIGLALQGSVPRTREPKVQQRLSEVIDDLQGVIQEI
TH_0388|M.thermoresistible__bu IARDLHDHVIQRLFAVGLTLQGTIPRARSTEVQQRLSECIDDLQAVIGEI
MAB_3890c|M.abscessus_ATCC_199 IARDLHDHVIQRLFAVGLAMQSTHRRSVDPTVAARLADHIDQLHEVIQEI
MAV_4108|M.avium_104 IAHDLHDHVIQRLFAAGMDLQGTLARARSPEVADRLNRTLDDLQTIIEEI
MLBr_00803|M.leprae_Br4923 TIREIHHRVKNNLQTVAALLRLQARRMANAEGREALIESVRRVSSIALVH
:::*.:* :.* : . :: * : : : :
Mflv_3844|M.gilvum_PYR-GCK RTAIFDLHGGSSAVTRLRQRLDAAVAAFPES-GVRTSMQCAGPLSVVDAG
Mvan_1395|M.vanbaalenii_PYR-1 RTAIFDLHGGHGGSNRLRQRLDEAIAQFTAS-DVHTTVQFSGPLSVVDPA
Mb2052c|M.bovis_AF2122/97 RSAIFDLHAGPSRATGLRHRLDKVIDQLAIP-ALHTTVQYTGPLSVVDTV
Rv2027c|M.tuberculosis_H37Rv RSAIFDLHAGPSRATGLRHRLDKVIDQLAIP-ALHTTVQYTGPLSVVDTV
MSMEG_5241|M.smegmatis_MC2_155 RTAIFDLHHTAPGGTRLRQRLDEAIAQFSGA-GLHTTSQFVGPLSVVDAE
MMAR_1517|M.marinum_M RTTIFDLHGSSQGVTRLRQRIDAAVAAFGSS-GLRTTVQFIGPLSVVDNV
MUL_2422|M.ulcerans_Agy99 RTTIFDLHGSSQGVTRLRQRIDAAVAAFGSS-GLRTTVQFIGPLSVVDNV
TH_0388|M.thermoresistible__bu RTTIFDLHGGPATGSQIRQRIGEAANQFSDA-GVRTRVQFAGPLSVISAV
MAB_3890c|M.abscessus_ATCC_199 RTAIFDLHTNDES---LRATLRSTINELTAETGLRVSVRMSGPLDVVPAS
MAV_4108|M.avium_104 RTTIFALRSPAAVAGDFRHRIQRVIAELTENRDLVTTVRMDGPMTAVGAE
MLBr_00803|M.leprae_Br4923 DALSMSVDEQVNLDEVIDRILPIMNDVASVD--RPIRINRVGNLGMLDSD
: : : : : . * : :
Mflv_3844|M.gilvum_PYR-GCK LADHAEAVVREAVSNAVRHAEASTVA------VTVTVEDDLAITVTDDGR
Mvan_1395|M.vanbaalenii_PYR-1 LSDHAEAVVREAVSNAIRHSGATTLD------VSVTAADELIIEVADDGC
Mb2052c|M.bovis_AF2122/97 LANHAEAVLREAVSNAVRHANATSLA------INVSVEDDVRVEVVDDGV
Rv2027c|M.tuberculosis_H37Rv LANHAEAVLREAVSNAVRHANATSLA------INVSVEDDVRVEVVDDGV
MSMEG_5241|M.smegmatis_MC2_155 LADHAEAVLREAVSNVVRHSGANELT------VRVKVEDDLSIEVTDDGE
MMAR_1517|M.marinum_M LADHAEAVVREAVSNAVRHAQATTLT------VRVKVDDDLCIEVSDNGR
MUL_2422|M.ulcerans_Agy99 LADHAEAVVREAVSNAVRHAQATTLT------VRVKVDDDLCIEVSDNGR
TH_0388|M.thermoresistible__bu LADHAEAVVREAVSNAVRHGGATDLS------VDVSVDDDLTIEVTDNGR
MAB_3890c|M.abscessus_ATCC_199 TSGDAEAVVREAVSNVVRHARARELS------VTVSVDDELVIEVVDDGI
MAV_4108|M.avium_104 LAEHAEAVTAEAVSNAVRHSGASRLT------VQIGVADMFTLDVIDNGR
MLBr_00803|M.leprae_Br4923 RATALIMVITELVQNAIEHAFDPASQEGFVAISAERSARWLDVVVHDDGR
: * * *.*.:.*. . : * *:*
Mflv_3844|M.gilvum_PYR-GCK GIA-----ADVTGSGLANLARRAQ-DVGGSFSVRR-GAQVGTTLTWRAPL
Mvan_1395|M.vanbaalenii_PYR-1 GMP-----ADVTESGLTNLRQRAG-DVGGTFTVES-GDAGGTRLRWCAPL
Mb2052c|M.bovis_AF2122/97 GIS-----GDITESGLRNLRQRAD-DAGGEFTVEN-MPTGGTLLRWSAPL
Rv2027c|M.tuberculosis_H37Rv GIS-----GDITESGLRNLRQRAD-DAGGEFTVEN-MPTGGTLLRWSAPL
MSMEG_5241|M.smegmatis_MC2_155 GIS-----GPVTESGLSNLRQRAD-QCGGELRIID-RPGGGTILRWTAPL
MMAR_1517|M.marinum_M GMP-----KTYTSSGLTNLRRRAQ-EAGGQFTIESPAAGGGTLLRWSAPL
MUL_2422|M.ulcerans_Agy99 GMP-----KTYTSSGLTNLRRRAQ-EAGGQFTIESPAAGGGTLLRWSAPL
TH_0388|M.thermoresistible__bu GFPE----GEVTESGLANLRHRAR-HAGGTLEVHN-RPDGGARLHWSVPL
MAB_3890c|M.abscessus_ATCC_199 GIP-----DVVARSGLSGLANRAA-QSGGTFSVSA-MDTGGTRLVWSAPL
MAV_4108|M.avium_104 GIAA----GNTRRSGLANMTRRAE-QLGGSCEISS-PPGGGTRVHWTAPL
MLBr_00803|M.leprae_Br4923 GLPQGFNLERSDSLGLQIVRTLVSSELDGSLGMRK-APNRGTDVVLRVPI
*:. ** : . . .* : *: : .*:
Mflv_3844|M.gilvum_PYR-GCK P-------
Mvan_1395|M.vanbaalenii_PYR-1 R-------
Mb2052c|M.bovis_AF2122/97 R-------
Rv2027c|M.tuberculosis_H37Rv R-------
MSMEG_5241|M.smegmatis_MC2_155 PD------
MMAR_1517|M.marinum_M IQ------
MUL_2422|M.ulcerans_Agy99 IQ------
TH_0388|M.thermoresistible__bu T-------
MAB_3890c|M.abscessus_ATCC_199 P-------
MAV_4108|M.avium_104 LDH-----
MLBr_00803|M.leprae_Br4923 GRRARLLL