For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KAIFITGAGSGMGREGATLFHANGWRVGAIDRNEDGLAALRVQLGAERLWARAVDVTDKAALEGALADFC AGNVGGGLDMMWNNAGIGEGGWFEDVPYEAAVRVVDVNFKAVLTGAYAALPYLKKAPGSLMFSTSSSSGT YGMPRIAVYSATKHAVKGLTEALSVEWQRHGVRVADVLPGLIDTAILTSTRQHSDEGPYTISAEQIRAAA PKKGMFRLMPSSSVAEAAWRAYQHPTRLHWYVPRSIRWIDRLKGVSPEFVRRHIAKSLATLEPKRK*
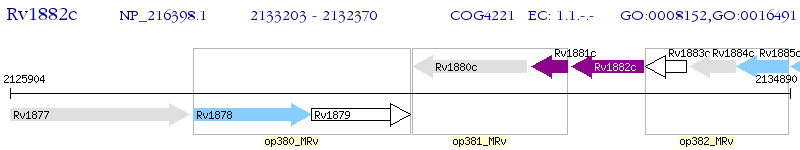
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1882c | - | - | 100% (277) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv3057c | - | 8e-20 | 31.61% (193) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1245c | - | 1e-18 | 35.79% (190) | short-chain type dehydrogenase/reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1914c | - | 1e-161 | 100.00% (277) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0407 | - | 5e-39 | 43.92% (189) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 8e-20 | 36.84% (190) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4335 | - | 3e-25 | 34.20% (193) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_2770 | - | 1e-131 | 83.03% (271) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2820 | - | 1e-132 | 80.87% (277) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3426 | - | 1e-20 | 32.35% (238) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1963 | - | 1e-100 | 65.96% (282) | PROBABLE SHORT-CHAIN TYPE DEHYDROGENASE/REDUCTASE |
| M. ulcerans Agy99 | MUL_2977 | - | 1e-130 | 83.03% (271) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0267 | - | 4e-37 | 43.09% (188) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0407|M.gilvum_PYR-GCK -------MTKSVFITGAATGIGRATALLFAGRGYRVGGYDLDEDGLAVLA
Mvan_0267|M.vanbaalenii_PYR-1 -------MTKSVFITGAATGIGRATALLFAQRGYRVGGYDIDEAGLRLLA
Rv1882c|M.tuberculosis_H37Rv --------MKAIFITGAGSGMGREGATLFHANGWRVGAIDRNEDGLAALR
Mb1914c|M.bovis_AF2122/97 --------MKAIFITGAGSGMGREGATLFHANGWRVGAIDRNEDGLAALR
MAV_2820|M.avium_104 --------MKSIFITGAGSGMGREGAKLFHAKGWRVGAVDRNDDGLATLQ
MMAR_2770|M.marinum_M --------MKSVFITGAGSGMGREGAKLFHAKGWRVAAVDRDGAGLDALG
MUL_2977|M.ulcerans_Agy99 --------MKSGFVTGAGSGMGREGAKLFHAKGWRVAAVDRDGAGLDALG
TH_1963|M.thermoresistible__bu ---VTRQTARAVFITGAGSGMGRAGAELFHARGWRVGAVDRNAEGLSGLR
MLBr_01094|M.leprae_Br4923 ---MEGFAGKVAVVTGAGSGIGQALAIELARSGAKLAISDVDGEGLAQTE
MAB_4335|M.abscessus_ATCC_1997 MGTAVQLAGKVVAVTGGARGIGRAIATAFAAEGAKVAIGDIDKKLCENTA
MSMEG_3426|M.smegmatis_MC2_155 --------MKTFFITGVSSGLGQAFANGALAAGHRVVGTVRKDSDAAAFE
: :** . *:*: * * :: .
Mflv_0407|M.gilvum_PYR-GCK ADITAVGGTPVVAHLDVTDPDEVARRLAEFAEG-SGGRIDVLINNAGLLN
Mvan_0267|M.vanbaalenii_PYR-1 ADITAVGGRPVVGRLDVTDADEVAHRLAEFVDA-AGGRLDVLINNAGLLQ
Rv1882c|M.tuberculosis_H37Rv VQLGAE--RLWARAVDVTDKAALEGALADFCAGNVGGGLDMMWNNAGIGE
Mb1914c|M.bovis_AF2122/97 VQLGAE--RLWARAVDVTDKAALEGALADFCAGNVGGGLDMMWNNAGIGE
MAV_2820|M.avium_104 QELGDD--RLWTRAVDVTDKAALDGALADFCAGNTGGGLDMMWNNAGIGE
MMAR_2770|M.marinum_M AELGTE--RLWTRVVDVTDRDALDRALADFCAGTSHGGLDMMWNNAGIGE
MUL_2977|M.ulcerans_Agy99 AELGTE--RLWTRVVDVTDRDALDRALADFCAGTSHGGLDMMWNNAGIGE
TH_1963|M.thermoresistible__bu AQLGPE--RLFTGQVDVTDRSALEAGLARFCADNTGGGLDMMWNNAGIGA
MLBr_01094|M.leprae_Br4923 GQLKAIGASARTDRLDVTEREAFLTYADVVHEN--FGKVNQIYNNAGIAF
MAB_4335|M.abscessus_ATCC_1997 AEIGNS---AIGLPLDVTDHGSFEAFFDTIAAT--VGPVDVIVNNAGIMP
MSMEG_3426|M.smegmatis_MC2_155 STAPDR---AHARQLDVTDDAAVTAVVAEVEKN--IGPIDVVIANAGYGV
:***: . . * :: : ***
Mflv_0407|M.gilvum_PYR-GCK AGRFEEIPLEVHHREIEVNVKGVVNGLHAAFPYLKGTPGAVVVNLASASA
Mvan_0267|M.vanbaalenii_PYR-1 AGRFEDIPLPVHHRAIEVNVKGVVNGLHAAFPYLRDTPDAVVVNLSSASA
Rv1882c|M.tuberculosis_H37Rv GGWFEDVPYEAAVRVVDVNFKAVLTGAYAALPYLKKAPGSLMFSTSSSSG
Mb1914c|M.bovis_AF2122/97 GGWFEDVPYEAAVRVVDVNFKAVLTGAYAALPYLKKAPGSLMFSTSSSSG
MAV_2820|M.avium_104 SGWFEDVPYDAAMRVVDVNYKAVLTGAYGALPYLKKSAGSLMFSTSSSSA
MMAR_2770|M.marinum_M GGWFEDVPYDAAMRVVDVNFKAVLTGAYGALPYLRKAPGSLMFSTSSSSA
MUL_2977|M.ulcerans_Agy99 GGWFEDVPYDAAMRVVDVNFKAVLTGAYGALPYLRKAPGSLMFSTSSSSA
TH_1963|M.thermoresistible__bu SGWFEDVPHEAALRVVAINFTAVLTGAYAALPYLRRTPGSLMFSTSSSSA
MLBr_01094|M.leprae_Br4923 TGDVEVSHFKDIERVMDVDYWGVVNGTKAFLPYLISSGDGHVINISSVFG
MAB_4335|M.abscessus_ATCC_1997 ITPFDDESLESIQRQLDINVRGVMWGSQLAIKRMKPRGGGVIVNIASAAG
MSMEG_3426|M.smegmatis_MC2_155 EGIFEETPIAEMRAQFATNVFGVAATLQAALPYLRERRKGHLMAVTSMGG
.: . : .* : : . :. :* .
Mflv_0407|M.gilvum_PYR-GCK IYGQAELANYSATKFFVRAITEALNLEWGHYG--IRVIDMWPLYVNTAMT
Mvan_0267|M.vanbaalenii_PYR-1 IYGQVELANYSATKFYVRAITEALNLEWAQHG--IRVIDMWPLYVNTAMT
Rv1882c|M.tuberculosis_H37Rv TYGMPRIAVYSATKHAVKGLTEALSVEWQRHG--VRVADVLPGLIDTAIL
Mb1914c|M.bovis_AF2122/97 TYGMPRIAVYSATKHAVKGLTEALSVEWQRHG--VRVADVLPGLIDTAIL
MAV_2820|M.avium_104 TYGMPRLAVYSSTKHAVKGLTEALSVEWQRHG--VRVADVLPGLIDTAIL
MMAR_2770|M.marinum_M TYGMPRLAVYSATKHAVKGLTEALSAEWRRHG--VRVADVLPGLIDTAIL
MUL_2977|M.ulcerans_Agy99 TYGMPRLAVYSATKHAVKGLTEALSAEWRRHG--VRVADVLPGLIDTAIL
TH_1963|M.thermoresistible__bu TYGMPQLAVYSATKHAVKGLTEALSVEWQRHG--VRVADIA---------
MLBr_01094|M.leprae_Br4923 LFSVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTTVYPGGIKTAIA
MAB_4335|M.abscessus_ATCC_1997 KAGFPGLATYCATKHAVVGLSESLSLEYEPAG--ISVVCVMPGMVNTELI
MSMEG_3426|M.smegmatis_MC2_155 LMAVPGMSAYCASKFAVEGMLESIRKEISGFG--IHVTAIEPGSFRTDWA
. : * ::*. * .: *:: * * : * :
Mflv_0407|M.gilvum_PYR-GCK KD------------------VKTGTTQSLGIRLTAQDIANDIVAAVDATW
Mvan_0267|M.vanbaalenii_PYR-1 QG------------------VKTGTTQSLGIRLTAQDIARDIVTAVEATW
Rv1882c|M.tuberculosis_H37Rv TSTRQHSDEGPY--TISAEQIRAAAPKKGMFRLMPSSSVAEAAWRAYQHP
Mb1914c|M.bovis_AF2122/97 TSTRQHSDEGPY--TISAEQIRAAAPKKGMFRLMPSSSVAEAAWRAYQHP
MAV_2820|M.avium_104 TTTTNHSNDGAA--PMTAEELRATAPKKGMLRLMPASSVAEVAWRAYNHP
MMAR_2770|M.marinum_M TSTRQHSDASAP--TLSSEQIRAAAPKKGMFRLLPASSVAEAAWQAYQHP
MUL_2977|M.ulcerans_Agy99 TSTRQHSDASAP--TLSSEQIRAAAPKKGMFRLLPASSVAEAAWHAYQHP
TH_1963|M.thermoresistible__bu ----------------------ARAPRRGMFRLLPPSSVAETAWAAYHDT
MLBr_01094|M.leprae_Br4923 RN--------------------ATAAEGLDVSKIASRFDTWVAHTSPQHA
MAB_4335|M.abscessus_ATCC_1997 SGLDTHWLLKTVE-PEDIANAVVKAVRRGVFPVFVPKMFGGMYRTMSVLP
MSMEG_3426|M.smegmatis_MC2_155 GRSMTRADRSIADYDELFEPIRAARLKASGNQLGNPVKAAEAVLAILDEP
. .
Mflv_0407|M.gilvum_PYR-GCK AR-----KALAQVHFPVG-LQAKALALGARFSPAWLTRLVNKGLAGS---
Mvan_0267|M.vanbaalenii_PYR-1 PR-----RLVKQVHFPVG-GQAKALALGSRFSPAWLTRLINKRLAGS---
Rv1882c|M.tuberculosis_H37Rv TR----------LHWYVP-RSIRWIDRLKGVSPEFVRRHIAKSLATLEPK
Mb1914c|M.bovis_AF2122/97 TR----------LHWYVP-RSIRWIDRLKGVSPEFVRRHIAKSLATLEPK
MAV_2820|M.avium_104 RR----------LHWYVP-RSIRLIDVFKGLSPEFVRRSIVKSLPALMPK
MMAR_2770|M.marinum_M TR----------LHWYVP-PSVRWIDRLKGVSPEFVRSRILKSLPTFGAG
MUL_2977|M.ulcerans_Agy99 TR----------LHWYVP-PSIRWIDRLKGVSPEFVRSRILKSLPTFGTG
TH_1963|M.thermoresistible__bu GRSRWGRTRAGRLHWYVP-DSLKWIDRLKGVSAEFVRGRIVKQLPALIEE
MLBr_01094|M.leprae_Br4923 ARIILKAVRKKKARVLVG-PDAKVANVVVRFSGGAGYQRLFAQVASRLIL
MAB_4335|M.abscessus_ATCC_1997 RS----------TGKFLT-RKLGIQDFMLNAHGSDARKAYEDRASHSGPS
MSMEG_3426|M.smegmatis_MC2_155 EP---------PAHLVLGSDALRLVGAARAAVDEDIRRWEELSRSTDFAD
: .
Mflv_0407|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0267|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1882c|M.tuberculosis_H37Rv RK------------------------------------------------
Mb1914c|M.bovis_AF2122/97 RK------------------------------------------------
MAV_2820|M.avium_104 RQ------------------------------------------------
MMAR_2770|M.marinum_M QQ------------------------------------------------
MUL_2977|M.ulcerans_Agy99 QQ------------------------------------------------
TH_1963|M.thermoresistible__bu DDRMKSPADIEGFEIDSRARQVYGEPHVTADGTTIVPVARVSRRGTGDAA
MLBr_01094|M.leprae_Br4923 NQR-----------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 TE------------------------------------------------
MSMEG_3426|M.smegmatis_MC2_155 GAQLAST-------------------------------------------
Mflv_0407|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0267|M.vanbaalenii_PYR-1 --------------------------------------------------
Rv1882c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1914c|M.bovis_AF2122/97 --------------------------------------------------
MAV_2820|M.avium_104 --------------------------------------------------
MMAR_2770|M.marinum_M --------------------------------------------------
MUL_2977|M.ulcerans_Agy99 --------------------------------------------------
TH_1963|M.thermoresistible__bu RVTARPVGVLVIKDGRATWKPTVDSTRIALFSQLIGLVAAALAGLAMVRR
MLBr_01094|M.leprae_Br4923 --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_3426|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0407|M.gilvum_PYR-GCK -------------------------
Mvan_0267|M.vanbaalenii_PYR-1 -------------------------
Rv1882c|M.tuberculosis_H37Rv -------------------------
Mb1914c|M.bovis_AF2122/97 -------------------------
MAV_2820|M.avium_104 -------------------------
MMAR_2770|M.marinum_M -------------------------
MUL_2977|M.ulcerans_Agy99 -------------------------
TH_1963|M.thermoresistible__bu PPWPDLHGDISSRPRPARPARSGRR
MLBr_01094|M.leprae_Br4923 -------------------------
MAB_4335|M.abscessus_ATCC_1997 -------------------------
MSMEG_3426|M.smegmatis_MC2_155 -------------------------