For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSPSVREWRDGGRWLPTAVGKVFVRSGPGDTPTMLLLHGYPSSSFDFRAVIPHLTGQAWVTMDFLGFGL SDKPRPHRYSLLEQAHLVETVVAHTVTGAVVVLAHDMGTSVTTELLARDLDGRLPFDLRRAVLSNGSVIL ERASLRPIQKVLRSPLGPVAARLVSRGGFTRGFGRIFSPAHPLSAQEAQAQWELLCYNDGNRIPHLLISY LDERIRHAQRWHGAVRDWPKPLGFVWGLDDPVATTNVLNGLRELRPSAAVVELPGLGHYPQVEAPKAYAE AALSLLVD
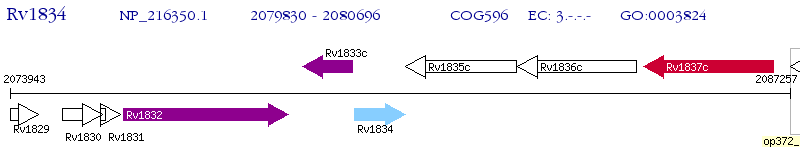
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1834 | - | - | 100% (288) | Probable hydrolase |
| M. tuberculosis H37Rv | Rv3176c | mesT | 8e-21 | 28.01% (307) | epoxide hydrolase MesT |
| M. tuberculosis H37Rv | Rv1833c | - | 6e-13 | 24.91% (277) | haloalkane dehalogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1865 | - | 1e-168 | 99.65% (288) | hydrolase |
| M. gilvum PYR-GCK | Mflv_2749 | - | 4e-19 | 28.52% (298) | alpha/beta hydrolase fold |
| M. leprae Br4923 | MLBr_02297 | - | 2e-06 | 23.91% (276) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_4300 | - | 4e-29 | 30.63% (284) | putative hydrolase, alpha/beta fold |
| M. marinum M | MMAR_2711 | - | 1e-140 | 82.93% (287) | hydrolase |
| M. avium 104 | MAV_2139 | - | 2e-23 | 31.67% (300) | hydrolase, alpha/beta fold family protein, putative |
| M. smegmatis MC2 155 | MSMEG_2777 | - | 1e-32 | 31.58% (285) | alpha/beta hydrolase fold |
| M. thermoresistible (build 8) | TH_4027 | - | 1e-107 | 65.12% (281) | PUTATIVE hydrolase |
| M. ulcerans Agy99 | MUL_3053 | - | 1e-136 | 80.84% (287) | hydrolase |
| M. vanbaalenii PYR-1 | Mvan_3788 | - | 9e-18 | 29.12% (261) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2749|M.gilvum_PYR-GCK ---------------MDIFDEWHRGGTTFSWTSTTPANGGAQVDVFARRT
Mvan_3788|M.vanbaalenii_PYR-1 ---------------MDIFTDWQRDGTHFTWTSTTAANAGTEVEVFSRRC
MAV_2139|M.avium_104 ---------------MDIFAQWQGGGTELRWRSTTAANDGREVGVFSRRC
Rv1834|M.tuberculosis_H37Rv ----------------------MTSPSVREWRDGGRWLPTAVGKVFVR-S
Mb1865|M.bovis_AF2122/97 ----------------------MTSPSVREWRDGGRWLPTAVGKVFVR-S
MMAR_2711|M.marinum_M ----------------------MTSPSVSDWRAGGGWINTAAGKVFVR-S
MUL_3053|M.ulcerans_Agy99 MNCRPITLVTLLSWSSRWYARRVTSPSVSDWRAGGGWINTAAGKVFVR-S
TH_4027|M.thermoresistible__bu ---------------------------VARWRAEGRWIRTGVGSVFVR-S
MAB_4300|M.abscessus_ATCC_1997 -----------------------MDAELEEWKAAGHYFDYLGFDIFYRVE
MSMEG_2777|M.smegmatis_MC2_155 -----------------------MHDELQRWKDAGQYFDYLGFDIFYRVD
MLBr_02297|M.leprae_Br4923 ----------------MAMSNPSVTRIAGPWHHLDVHANGIRFHVVEAVP
* :.
Mflv_2749|M.gilvum_PYR-GCK GT----------PGAPALVCVHGFPTAGVDYHALTRELADDFDIYVLDFP
Mvan_3788|M.vanbaalenii_PYR-1 GT----------PGAPALVCVHGFPTSGIDYSALARELAAEFDIYVLDFP
MAV_2139|M.avium_104 GT----------PGAPALVLVHGFPTSSIDYFALAGELSRDFDIHLIDFP
Rv1834|M.tuberculosis_H37Rv GP----------GDTPTMLLLHGYPSSSFDFRAVIPHLT-GQAWVTMDFL
Mb1865|M.bovis_AF2122/97 GP----------GDTPTMLLLHGYPSSSFDFRAVIPHLT-GQAWVTMDFL
MMAR_2711|M.marinum_M GE----------GIAPTVLLLHGFPSSSFDFRSVIPYLA-GQAWLTMDFL
MUL_3053|M.ulcerans_Agy99 GE----------GIAPTVLLLHGFPSSSFNFRSVIPYLA-GQAWLTMDFL
TH_4027|M.thermoresistible__bu AS----------GRGSPVVLLHGYPSSSYDYRAVLPELG-DRGWVTFDFL
MAB_4300|M.abscessus_ATCC_1997 ------------GSGPPLLLIHGYPFNSWDWALIWPTLVERFTVIAPDMI
MSMEG_2777|M.smegmatis_MC2_155 GVAP--------GTGPNLLLIHGYPFNSWDWSLIWPTLTQRFTVIAPDMI
MLBr_02297|M.leprae_Br4923 TQPAGQDCAPSATARPLVMLLHGFGSFWWSWRHQLRGLT-EARLVAVDLR
. :: :**: .: * *:
Mflv_2749|M.gilvum_PYR-GCK GYGLSGKPPHPYVYSLYDDARLLVHAITDVWGLETFRMVTHDRGSSIGMI
Mvan_3788|M.vanbaalenii_PYR-1 GYGLSGKPPSPYVYSLYDDARLLVHAVTERWRLTDYRMITHDRGSSVGMI
MAV_2139|M.avium_104 GYGLSDKPPEPYVYSLYDDARLLVHAITEVWRLSDYRMLTHDRGSSVGMI
Rv1834|M.tuberculosis_H37Rv GFGLSDKP-RPHRYSLLEQAHLVETVVAHTV-TGAVVVLAHDMGTSVTTE
Mb1865|M.bovis_AF2122/97 GFGLSDKP-RPHRYSLLEQAHLVETVVAHTV-TGAVVVLAHDMGTSVTTE
MMAR_2711|M.marinum_M GFGLSDKP-RPHRYSLLEQADLVQNVVAQNV-TGPVVVLAHDMGTSVTTE
MUL_3053|M.ulcerans_Agy99 GFGLSDKP-RPHRYSLLEQADLVQNVVAQNV-TGPVVVLAHDMGTSVTTE
TH_4027|M.thermoresistible__bu GFGLSDKP-RPHRYSLFEQADLVQTVVRSVT-DEPVLLIAHDMGTSVATE
MAB_4300|M.abscessus_ATCC_1997 GMGFSDKP-VQYGYSVLDHADMHEALLSYLG-IERFHLLTHDLGNSVGQE
MSMEG_2777|M.smegmatis_MC2_155 GMGFSDKP-VAYEYRVTDHADIHEALLAHLG-ITSTHILAHDLGDSVGQE
MLBr_02297|M.leprae_Br4923 GYGGSDKP--PRGYDGWTLAGDTAGLIRALG-HSSATLVGHAEGGLACWA
* * *.** * * : :: * *
Mflv_2749|M.gilvum_PYR-GCK ALGLLDDIG-----RRPLDLFMTNANIYLPLANLTAFQVALLDDDTGRAT
Mvan_3788|M.vanbaalenii_PYR-1 ALGMLAESG-----STPVDVFITNGNIYLPLANLTAFQVALLDEATGRAT
MAV_2139|M.avium_104 ASGMLAALDPP---AAPVDLILTNANIYLPLANLTAFQSALLDPSTARAT
Rv1834|M.tuberculosis_H37Rv LLARDLDGRLP---FDLRRAVLSNGSVILERASLRPIQKVLRSP-LGPVA
Mb1865|M.bovis_AF2122/97 LLARDLDGRLP---FDLRRAVLSNGSVILERASLRPIQKVLRSP-LGPVA
MMAR_2711|M.marinum_M LLARDLEGRLP---FELQRAVLSNGSVILERASLRPIQKVLRGP-LGPIA
MUL_3053|M.ulcerans_Agy99 LLARDLEGRLP---FELQRVVLSNGSVILERASLRLIQKVLRGP-LGPIA
TH_4027|M.thermoresistible__bu LMARDLQGQLP---FRLQRVVLSNGSVLLDRASLRPIQKVLRGR-FGPVV
MAB_4300|M.abscessus_ATCC_1997 MLARFEFEQQSRGRVPIDSVTWLNGGLFIEAYRPRIAQTLMSRTPLGDLV
MSMEG_2777|M.smegmatis_MC2_155 MLARHEFGGQAYGGFQIESVTWLNGGLFNEAYRPRLMQKVMSRTPLGDIL
MLBr_02297|M.leprae_Br4923 TALLHPRLVRAIALINSPHPAALRRSMLTHRNQGYALLPTLLRYQLPIWP
. .: :
Mflv_2749|M.gilvum_PYR-GCK AAATTP---EMLAAGLGAA---TFMPRRTLQDPEIAALAHCFAHNDGIRV
Mvan_3788|M.vanbaalenii_PYR-1 AAATTP---AMLAAGLGAS---TFMPRRSPQDDEIAALAQCFAHNDGISV
MAV_2139|M.avium_104 AAATTP---ELLAAGMGAS---TFMPRRTLDDPEIAALAKCFAHNDGIRV
Rv1834|M.tuberculosis_H37Rv ARLVSR---GGFTRGFGR----IFSPAHPLSAQEAQAQWELLCYNDGNRI
Mb1865|M.bovis_AF2122/97 ARLVSR---GGFTRGFGR----IFSPAHPLSAQEAQAQWELLCYNDGNRI
MMAR_2711|M.marinum_M ARLVNR---GGFTRGFGK----LFSAQHPLSAQEAEAQWELLTHNNGHRI
MUL_3053|M.ulcerans_Agy99 ARLVNR---GGFTRGFGK----LFSAQHPLSAQEPEAQWELLTHNNGHRI
TH_4027|M.thermoresistible__bu SQLVNE---PMFIRGLGK----VFSKQRPLSAAEGAAQWALMSYHDGHRI
MAB_4300|M.abscessus_ATCC_1997 SRFQGT---PVTRRIMDAAVSEMFGPDTKPSPRLLSLFQQVLEYNDGARV
MSMEG_2777|M.smegmatis_MC2_155 SPLQGS---PLSRRVFESTLDQMFGVNTKPSREMIEKFHQILDHNDGKRV
MLBr_02297|M.leprae_Br4923 ERLLTRNNAAEIERLVRSRSSAKWQACEDFAVTIDHLRIAIQIPAAAHCA
. : .
Mflv_2749|M.gilvum_PYR-GCK LPDTVRYLHERAADETG--WLERLSTIPVDTTVIWGLHDAVAPLRVANHV
Mvan_3788|M.vanbaalenii_PYR-1 LPDTVRYLHERAADETG--WLERLSTIDVDTTLIWGLHDTVAPLRVANHV
MAV_2139|M.avium_104 LPDTIQYLNERAADETR--WLEALSTSDVDTTMVWGLHDNVAPLRVPNHV
Rv1834|M.tuberculosis_H37Rv PHLLISYLDERIRHAQR--WHGAVRDWPKPLGFVWGLDDPVATTNVLNGL
Mb1865|M.bovis_AF2122/97 PHLLISYLDERIRHAQR--WHGAVRDWPKPLGFVWGLDDPVATTNVLNGL
MMAR_2711|M.marinum_M AHLLISYLEERERYAQR--WHNAVRDWPKPLGFVWGLDDPVATTNVLNGL
MUL_3053|M.ulcerans_Agy99 AHLLISYLEERERYAQR--WNNAVRDWPKPLGFVWGLDDPVATTNVLNGL
TH_4027|M.thermoresistible__bu MHLLTAYLNERVRYADR--WHGAVADWPKPLGLLWGTDDPVATTDVLAGL
MAB_4300|M.abscessus_ATCC_1997 THQVGRFINDRYDNRSR--WVRAMRETAVPMRMIDGPIDPNSGLHMAERY
MSMEG_2777|M.smegmatis_MC2_155 LHKVGRFVTDRYTHRNR--WVRAMRETSVPMRLIDGPSDPNSGRHMAQRY
MLBr_02297|M.leprae_Br4923 LEYQRWAVRSQLRNEGRKFMKSMSRPFSVPLLHVRGDADPYLLADSVDRT
: .: : * *
Mflv_2749|M.gilvum_PYR-GCK WQTYVKPKPGRNHYWVVPTADHYVQCDAPAELAQIIRMTADGADIDLQTL
Mvan_3788|M.vanbaalenii_PYR-1 WEHYLKSSPGRNRYWVVPGADHYVQCDAPGQLAGIIRLTAEGVPPGLPTL
MAV_2139|M.avium_104 WQTYLKTKPGRNRYWVIPGADHYLQCDAPAQLAEIVRLTARGEDVPLQTL
Rv1834|M.tuberculosis_H37Rv RELRPS-----AAVVELPGLGHYPQVEAPKAYAEAALSLL---------V
Mb1865|M.bovis_AF2122/97 RELRPS-----AAVVELPGLGHYPQAEAPKAYAEAALSLL---------V
MMAR_2711|M.marinum_M RELRPN-----AGVIELPGLGHYPQVEDPKAYTQAALDLL---------V
MUL_3053|M.ulcerans_Agy99 RELRPN-----AGVIELPGLGHYPQVEDPKAYTQPALDLL---------V
TH_4027|M.thermoresistible__bu RELRSD-----AEVIELAGVGHYPQIEVPEVFARAALELLGLN----RTI
MAB_4300|M.abscessus_ATCC_1997 LEVIHDP----DVVLLDDNIGHWPQIEAPEAVLTHFLEHIDRITP-----
MSMEG_2777|M.smegmatis_MC2_155 LEVIPDP----DVVMLADDIAHWPQIEAPDAVLAHFLEHIDRVVAL----
MLBr_02297|M.leprae_Br4923 RRYAPHG-----RYVSVSDAGHFSHEEAPEEINRYLTRFLKQVHGPRLS-
. *: : : *
Mflv_2749|M.gilvum_PYR-GCK GNRPDGAVLVDRSDGDRL
Mvan_3788|M.vanbaalenii_PYR-1 GDRPDGAVLVDRSDGDDG
MAV_2139|M.avium_104 GDRPDGAVLVDGGAS---
Rv1834|M.tuberculosis_H37Rv D-----------------
Mb1865|M.bovis_AF2122/97 D-----------------
MMAR_2711|M.marinum_M G-----------------
MUL_3053|M.ulcerans_Agy99 G-----------------
TH_4027|M.thermoresistible__bu G-----------------
MAB_4300|M.abscessus_ATCC_1997 ------------------
MSMEG_2777|M.smegmatis_MC2_155 ------------------
MLBr_02297|M.leprae_Br4923 ------------------