For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SIDFTPDPQLYPFESRWFDSSRGRIHYVDEGTGPPILLCHGNPTWSFLYRDIIVALRDRFRCVAPDYLGF GLSERPSGFGYQIDEHARVIGEFVDHLGLDRYLSMGQDWGGPISMAVAVERADRVRGVVLGNTWFWPADT LAMKAFSRVMSSPPVQYAILRRNFFVERLIPAGTEHRPSSAVMAHYRAVQPNAAARRGVAEMPKQILAAR PLLARLAREVPATLGTKPTLLIWGMKDVAFRPKTIIPRLSATFPDHVLVELPNAKHFIQEDAPDRIAAAI IERFG*
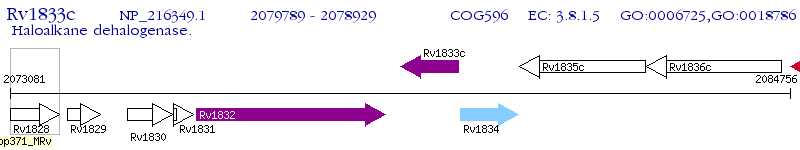
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1833c | - | - | 100% (286) | haloalkane dehalogenase |
| M. tuberculosis H37Rv | Rv2296 | - | 4e-26 | 31.17% (308) | haloalkane dehalogenase |
| M. tuberculosis H37Rv | Rv2579 | dhaA | 6e-21 | 29.47% (285) | haloalkane dehalogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1864c | - | 1e-169 | 100.00% (286) | haloalkane dehalogenase |
| M. gilvum PYR-GCK | Mflv_1960 | - | 3e-16 | 25.10% (255) | alpha/beta hydrolase fold |
| M. leprae Br4923 | MLBr_00862 | ephD | 2e-05 | 31.91% (94) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2289 | - | 5e-21 | 31.16% (276) | putative hydrolase, alpha/beta hydrolase fold |
| M. marinum M | MMAR_2710 | - | 1e-142 | 83.04% (283) | haloalkane dehalogenase |
| M. avium 104 | MAV_2130 | - | 2e-29 | 32.89% (304) | haloalkane dehalogenase |
| M. smegmatis MC2 155 | MSMEG_6837 | - | 8e-20 | 30.00% (270) | haloalkane dehalogenase |
| M. thermoresistible (build 8) | TH_1615 | linB | 2e-22 | 29.23% (284) | POSSIBLE HALOALKANE DEHALOGENASE DHAA (1-CHLOROHEXANE |
| M. ulcerans Agy99 | MUL_3052 | - | 1e-140 | 81.98% (283) | haloalkane dehalogenase |
| M. vanbaalenii PYR-1 | Mvan_2384 | - | 3e-21 | 29.43% (299) | haloalkane dehalogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1833c|M.tuberculosis_H37Rv ---------MSIDFTPDP-----QLYPFESRWFDSSRG-----RIHYVDE
Mb1864c|M.bovis_AF2122/97 ---------MSIDFTPDP-----QLYPFESRWFDSSRG-----RIHYVDE
MMAR_2710|M.marinum_M -----MGRQFTPDFTPDP-----KLYPFASRWFDSSRG-----RLHYIDE
MUL_3052|M.ulcerans_Agy99 -----MGRQFTPDFTPDP-----KLYPFASRWFDSSRG-----RLHYIDE
MAV_2130|M.avium_104 MPVHAEELNMHVLRTPDSRFENLEDYPFVAHYLDVTARDTRPLRMHYLDE
Mvan_2384|M.vanbaalenii_PYR-1 ---------MDILRTPDERFADLPDFPFAPHYVEVDG-----LRMHYLDE
MSMEG_6837|M.smegmatis_MC2_155 --------------MPGS-------EPYGRLQYREING----KRMAYIDE
TH_1615|M.thermoresistible__bu --------------MPGS-------KPYGEPRFREIKN----KRMAYIDE
Mflv_1960|M.gilvum_PYR-GCK ---------------------------------MTVRG----VDFHITEA
MLBr_00862|M.leprae_Br4923 --------------MPAT----QSIHKVPHQFVESPDG----VRLAIYQS
MAB_2289|M.abscessus_ATCC_1997 --MGVAAHQQIYRPVSGHGGAMSLGLEKHRRSVDIERG-----RISYLDI
: :
Rv1833c|M.tuberculosis_H37Rv G--TGPPILLCHGNPTWSFLYRDIIVALRDR-FRCVAPDYLGFGLSERPS
Mb1864c|M.bovis_AF2122/97 G--TGPPILLCHGNPTWSFLYRDIIVALRDR-FRCVAPDYLGFGLSERPS
MMAR_2710|M.marinum_M G--DGPPILLCHGNPTWSFLYRHIIVALRDH-FRCIAPDYLGFGLSEHPG
MUL_3052|M.ulcerans_Agy99 S--DGPPILLCHGNPTWSFLYRHIIVALRDH-FRCIAPDYLGFGLSEHPG
MAV_2130|M.avium_104 GPIDGPPIVLLHGEPTWSYLYRTMITPLTDAGNRVLAPDLIGFGRSDKPS
Mvan_2384|M.vanbaalenii_PYR-1 GPADGPVVLLLHGEPSWSYLYRRMIPVLVDAGLRAVAIDLVGFGRSDKPT
MSMEG_6837|M.smegmatis_MC2_155 A--RGDAIVFQHGNPSSSYLWRNVLPHTEGL-GRLVACDLIGMGASDKLD
TH_1615|M.thermoresistible__bu G--EGDAIVFQHGNPTSSYLWRNVLPHLEGL-GRLVACDLIGMGGSEKLS
Mflv_1960|M.gilvum_PYR-GCK GPEDGRPVLALHGWPQHHWAYRDLLGDPPPG-LRIIAPDLPGYGWSG-PP
MLBr_00862|M.leprae_Br4923 GQPEGPAIVLVHGFPDSHVLWDGVVPLLAKR-FRIVRYDNRGVGQSSVPK
MAB_2289|M.abscessus_ATCC_1997 G--AGSPTVFVHGVLTNSLLWRDVVTAVAATGRRCIALDLPGHGHSPVPA
. * : ** : :: * : * * * *
Rv1833c|M.tuberculosis_H37Rv GFG---YQIDEHARVIGEFVDHLGLDR-YLSMGQDWG--GPISMAVAVER
Mb1864c|M.bovis_AF2122/97 GFG---YQIDEHARVIGEFVDHLGLDR-YLSMGQDWG--GPISMAVAVER
MMAR_2710|M.marinum_M TFG---YKVDEHAQVVGEFVDHLGLDN-YLTMGQDWG--GPISMAVAVER
MUL_3052|M.ulcerans_Agy99 TFG---YKVDEHAQVVGEFVDHLGLDN-YLTMGQDWG--GPISMAVAVER
MAV_2130|M.avium_104 RIED--YSYQRHVDWVVSWFEHLNLSD-VTLFVQDWG--SLIGLRIAAEQ
Mvan_2384|M.vanbaalenii_PYR-1 RRED--YTYQAHVDWTWAAIEAIGLTG-ITLVCQDWG--GLIGLRLVGEH
MSMEG_6837|M.smegmatis_MC2_155 GSGPDSYHYHENRDYLFALWDALDLGDRVTLVLHDWG--GALGFDWANRH
TH_1615|M.thermoresistible__bu PSGPDRYHYGEHRDYLFALWEELELGDRVVLVLHDWG--SVLGFDWANHH
Mflv_1960|M.gilvum_PYR-GCK PHR---WAKEDVVTDLVALVDALGLDR-FLLVGHDWG--GYIGHLLALRI
MLBr_00862|M.leprae_Br4923 PVS--AYTMTRFADDFAAVIDELSPDRPVHVLAHDWGSVGVWEYLSRSGA
MAB_2289|M.abscessus_ATCC_1997 DGID--VTLSGLAQLIAEFLDSIGIDE-FDLVANDTG--GAVAQIMVARQ
: : . :* * .
Rv1833c|M.tuberculosis_H37Rv ADRVRGVVLGN----------------------TWFWPADTL----AMKA
Mb1864c|M.bovis_AF2122/97 ADRVRGVVLGN----------------------TWFWPADTL----AMKA
MMAR_2710|M.marinum_M AERVRGIVLGN----------------------TWFWPADTL----TMKA
MUL_3052|M.ulcerans_Agy99 AERVRGIVLGN----------------------TWFWPADTL----TMKA
MAV_2130|M.avium_104 PDRVGRLVVAN----------------------GFLPTAQRR----TPPA
Mvan_2384|M.vanbaalenii_PYR-1 PDRFARVVAAN----------------------TTLPTGDQR----PGEA
MSMEG_6837|M.smegmatis_MC2_155 RDRVAGIVHME----------------------TVSVPMEWDDFPDEVAQ
TH_1615|M.thermoresistible__bu RDRVQGIAHME----------------------SIVTPLTWADWEDPARS
Mflv_1960|M.gilvum_PYR-GCK PERIDAYLALN----------------------IAHPWVPPKVMAPHLWR
MLBr_00862|M.leprae_Br4923 SDRVASFTSVSGPSQDQLVDYILSGLRRPWRPRTFARAVSQLFRLTYMVL
MAB_2289|M.abscessus_ATCC_1997 PDRIRTLALTN----------------------CDTEGHVPPKLFKPVVA
:*. .
Rv1833c|M.tuberculosis_H37Rv FSRVMSSPPVQYAILRR---------NFFVERLIPAGTEH--RPSSAVMA
Mb1864c|M.bovis_AF2122/97 FSRVMSSPPVQYAILRR---------NFFVERLIPAGTEH--RPSSAVMA
MMAR_2710|M.marinum_M FSTVMSSLPMQYAILHH---------NFFVERLIPAGTTH--RPSSAVME
MUL_3052|M.ulcerans_Agy99 FSTVMSSLPMQYAILHH---------NFFVERLIPAGTTH--RPSSAVME
MAV_2130|M.avium_104 FYAWR-----AFARYSP---------VLPAGRIVSVGTVR--RVSSKVRA
Mvan_2384|M.vanbaalenii_PYR-1 FLAWQ-----RFSQETP---------DFAVGHIVNGGCVS--TLAPEVIA
MSMEG_6837|M.smegmatis_MC2_155 MFRGLRSPQGEEMVLEN---------NAFIEGVLPSIVMR--TLSEEEMI
TH_1615|M.thermoresistible__bu VFQGLRTPEGERMVLEE---------NIFVEAMLPRGVHR--TLSDEEMA
Mflv_1960|M.gilvum_PYR-GCK FLLYQPVVASAGVPLQTR--------TRYLEKVVFRAAAP--KLDAATVG
MLBr_00862|M.leprae_Br4923 FSIPVLVPMVLWIALSSATIRRTMVDNISTDQIHHSATLA--RDAAHSVK
MAB_2289|M.abscessus_ATCC_1997 LARPPLMALIGPRLFAR---------RKVLRALLGAGYRKPGRLPDSIVE
. :
Rv1833c|M.tuberculosis_H37Rv HYRAVQPNA-AARRG-----------------------------------
Mb1864c|M.bovis_AF2122/97 HYRAVQPNA-AARRG-----------------------------------
MMAR_2710|M.marinum_M HYRGVQPTA-SARVG-----------------------------------
MUL_3052|M.ulcerans_Agy99 HYRGVQPTA-SARVG-----------------------------------
MAV_2130|M.avium_104 GYDAPFPDK-TYQAG-----------------------------------
Mvan_2384|M.vanbaalenii_PYR-1 AYDAPFPDD-SYKAG-----------------------------------
MSMEG_6837|M.smegmatis_MC2_155 HYRRPFLNAGEDRRP-----------------------------------
TH_1615|M.thermoresistible__bu HYREPFRDRGEGRRP-----------------------------------
Mflv_1960|M.gilvum_PYR-GCK TYAERFRDPVVARTA-----------------------------------
MLBr_00862|M.leprae_Br4923 TYPANYFRAFSGRRRGKGVPVVNVPVQLIVNTMDRYVRPYGYDATARWVP
MAB_2289|M.abscessus_ATCC_1997 SYGQPVFGTAESSRF-----------------------------------
*
Rv1833c|M.tuberculosis_H37Rv -------------VAEMPKQILAARPLLARLAR--EVPATLG---TKPTL
Mb1864c|M.bovis_AF2122/97 -------------VAEMPKQILAARPLLARLAR--EVPATLG---TKPTL
MMAR_2710|M.marinum_M -------------VAEMPKQILAARPLLARLAS--EVPAQLG---TKPAL
MUL_3052|M.ulcerans_Agy99 -------------VAEMPKQILAARPLLARLAS--EVPAQLG---TKPAL
MAV_2130|M.avium_104 -------------ARAFPQLVPTSPADPAIPAN--RKAWEALGRWEKPFL
Mvan_2384|M.vanbaalenii_PYR-1 -------------ARQFPTLVPTSPDDPAAPAN--RAAWEVLRTFDRPFL
MSMEG_6837|M.smegmatis_MC2_155 -------------TLSWPRDVPLA-GEPAEVVA--VIEDFGEWLATSDIP
TH_1615|M.thermoresistible__bu -------------TLTWPRNVPID-GEPADVAA--IVADYAVWLAEGDVP
Mflv_1960|M.gilvum_PYR-GCK -------------RDTYRTFLTREIPKGVRDGE--RRWSTVP------TR
MLBr_00862|M.leprae_Br4923 RLWRRDIKAGHFSPMSHPQVMAAAVHDLADLVDGKQPSRALLRAQVGRSR
MAB_2289|M.abscessus_ATCC_1997 ----------------LSKMLRAMHTRDLEAAR------AGLVTFTKPTL
:
Rv1833c|M.tuberculosis_H37Rv LIWGMKDV-----------------AFR-PKTIIPRLSATFPDHVLVELP
Mb1864c|M.bovis_AF2122/97 LIWGMKDV-----------------AFR-PKTIIPRLSATFPDHVLVELP
MMAR_2710|M.marinum_M LVWGMKDF-----------------AFR-PEPMLARMRAVFADHVLVELP
MUL_3052|M.ulcerans_Agy99 LVWGMKDF-----------------AFR-PEPMLARMRAVFADHILVELP
MAV_2130|M.avium_104 AIFGARDP-----------------ILGHADSPLIKHIPGAAGQPHARIN
Mvan_2384|M.vanbaalenii_PYR-1 CAFSDSDP-----------------ITRGADAPLRKFIPGAQGRPEVTIA
MSMEG_6837|M.smegmatis_MC2_155 KLFIRADP-----------------GVIQGKQRILDIVRSWPNQTEITVP
TH_1615|M.thermoresistible__bu KLFVNAEP-----------------GAIVRG-RVRDFVRSWPNQTEITVA
Mflv_1960|M.gilvum_PYR-GCK VLFGRSDT-----------------AIH---PDLAAAETARADDYTIDVV
MLBr_00862|M.leprae_Br4923 DTFGDTLVSVTGAGSGIGRETALALARRGAEVVVSDIDEAAAKNTAAQIV
MAB_2289|M.abscessus_ATCC_1997 IAWGLGDY------------------FFPLKYGQRLAELFGGPVTFETVP
: :
Rv1833c|M.tuberculosis_H37Rv NAKHFIQEDAPDRIAAAIIERFG---------------------------
Mb1864c|M.bovis_AF2122/97 NAKHFIQEDAPDRIAAAIIERFG---------------------------
MMAR_2710|M.marinum_M EAKHFIQEDAPDEIAAAIIARFG---------------------------
MUL_3052|M.ulcerans_Agy99 EAKHLIQEDAPDEIAAAIIARFG---------------------------
MAV_2130|M.avium_104 -ASHFIQEDRGPELAERILSWQQALL------------------------
Mvan_2384|M.vanbaalenii_PYR-1 GGGHFLQEDKGPELAAVVVDFIAANPV-----------------------
MSMEG_6837|M.smegmatis_MC2_155 -GTHFLQEDSADQIGEAIASFVREIRAGDNLHREAAPEMNSAS-------
TH_1615|M.thermoresistible__bu -GRHYIQEDSPDEIGAAIADFVRKLRG-----------------------
Mflv_1960|M.gilvum_PYR-GCK DGGHFILDEQPDLVRAKLIAVADEFPERPVR-------------------
MLBr_00862|M.leprae_Br4923 ASGGVAYAYALDVSDAAAVEAFAEQVSAKHGVPDIVVNNAGIGQAGRFLD
MAB_2289|M.abscessus_ATCC_1997 DGRLYFPDEDADRLLPLLYRHWHSAGRQESSTEVGS--------------
.
Rv1833c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1864c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2710|M.marinum_M --------------------------------------------------
MUL_3052|M.ulcerans_Agy99 --------------------------------------------------
MAV_2130|M.avium_104 --------------------------------------------------
Mvan_2384|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6837|M.smegmatis_MC2_155 --------------------------------------------------
TH_1615|M.thermoresistible__bu --------------------------------------------------
Mflv_1960|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 TPPEEFDRVLAVNLGGVVNCCRAFGQRLVERGTGGHIVNVSSMAAYAPLQ
MAB_2289|M.abscessus_ATCC_1997 --------------------------------------------------
Rv1833c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1864c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2710|M.marinum_M --------------------------------------------------
MUL_3052|M.ulcerans_Agy99 --------------------------------------------------
MAV_2130|M.avium_104 --------------------------------------------------
Mvan_2384|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6837|M.smegmatis_MC2_155 --------------------------------------------------
TH_1615|M.thermoresistible__bu --------------------------------------------------
Mflv_1960|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 SLSAYCTSKAATYMFSDCLRAELDAVGVGLTTICPGVIDTNIIQSTRIDT
MAB_2289|M.abscessus_ATCC_1997 --------------------------------------------------
Rv1833c|M.tuberculosis_H37Rv --------------------------------------------------
Mb1864c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_2710|M.marinum_M --------------------------------------------------
MUL_3052|M.ulcerans_Agy99 --------------------------------------------------
MAV_2130|M.avium_104 --------------------------------------------------
Mvan_2384|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6837|M.smegmatis_MC2_155 --------------------------------------------------
TH_1615|M.thermoresistible__bu --------------------------------------------------
Mflv_1960|M.gilvum_PYR-GCK --------------------------------------------------
MLBr_00862|M.leprae_Br4923 PTAKQERVRDRRGQLDMMFRLRRYGPDKVADTILSAIKKNKPIRPVAPEA
MAB_2289|M.abscessus_ATCC_1997 --------------------------------------------------
Rv1833c|M.tuberculosis_H37Rv -----------------------
Mb1864c|M.bovis_AF2122/97 -----------------------
MMAR_2710|M.marinum_M -----------------------
MUL_3052|M.ulcerans_Agy99 -----------------------
MAV_2130|M.avium_104 -----------------------
Mvan_2384|M.vanbaalenii_PYR-1 -----------------------
MSMEG_6837|M.smegmatis_MC2_155 -----------------------
TH_1615|M.thermoresistible__bu -----------------------
Mflv_1960|M.gilvum_PYR-GCK -----------------------
MLBr_00862|M.leprae_Br4923 YALYGISRVLPQVLRSTARIRVI
MAB_2289|M.abscessus_ATCC_1997 -----------------------