For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPAPSAEVFDRLRNLAAIKDVAARPTRTIDEVFTGKPLTTIPVGTAADVEAAFAEARAAQTDWAKRPVIE RAAVIRRYRDLVIENREFLMDLLQAEAGKARWAAQEEIVDLIANANYYARVCVDLLKPRKAQPLLPGIGK TTVCYQPKGVVGVISPWNYPMTLTVSDSVPALVAGNAVVLKPDSQTPYCALACAELLYRAGLPRALYAIV PGPGSVVGTAITDNCDYLMFTGSSATGSRLAEHAGRRLIGFSAELGGKNPMIVARGANLDKVAKAATRAC FSNAGQLCISIERIYVEKDIAEEFTRKFGDAVRNMKLGTAYDFSVDMGSLISEAQLKTVSGHVDDATAKG AKVIAGGKARPDIGPLFYEPTVLTNVAPEMECAANETFGPVVSIYPVADVDEAVEKANDTDYGLNASVWA GSTAEGQRIAARLRSGTVNVDEGYAFAWGSLSAPMGGMGLSGVGRRHGPEGLLKYTESQTIATARVFNLD PPFGIPATVWQKSLLPIVRTVMKLPGRR
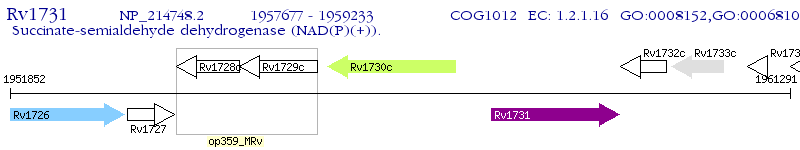
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1731 | gabD2 | - | 100% (518) | succinic semialdehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0223c | - | 2e-64 | 34.95% (455) | aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv0234c | gabD1 | 2e-59 | 32.11% (464) | succinic semialdehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1760 | gabD2 | 0.0 | 100.00% (518) | succinic semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0132 | - | 4e-80 | 39.96% (463) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 4e-60 | 32.07% (449) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4136c | - | 7e-79 | 37.89% (475) | putative aldehyde dehydrogenase |
| M. marinum M | MMAR_2571 | gabD2 | 0.0 | 90.93% (518) | succinate-semialdehyde dehydrogenase |
| M. avium 104 | MAV_4946 | - | 1e-67 | 35.19% (466) | aldehyde dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_5912 | gabD2 | 0.0 | 74.27% (517) | succinic semialdehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_3138 | - | 7e-82 | 40.08% (474) | PUTATIVE aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_3190 | gabD2 | 0.0 | 90.15% (518) | succinic semialdehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0771 | - | 4e-82 | 40.56% (466) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0132|M.gilvum_PYR-GCK ---MTATSDAHTANPF----------TGTGTITNPATGAVAGTVHWTDPA
Mvan_0771|M.vanbaalenii_PYR-1 MGFMTATSDVQPPHTF----------TGTGTIRNPATGAVAGEVRWTDPA
TH_3138|M.thermoresistible__bu ---MTATSGVDA--------------TETATIRNPATGGVAGEVRWTDPA
MAB_4136c|M.abscessus_ATCC_199 ---MTTTSENLVSTPS----------DTSHDVRNPANGQVVGRVNWTDPS
Rv1731|M.tuberculosis_H37Rv ---MPAPSAEVFDRLRNLAAIKDVAARPTRTIDEVFTGKPLTTIPVGTAA
Mb1760|M.bovis_AF2122/97 ---MPAPSAEVFDRLRNLAAIKDVAARPTRTIDEVFTGKPLTTIPVGTAA
MMAR_2571|M.marinum_M ---MPVPSSAVFERLLSLAAIKDVDARPTRTIDEVFTGKPLTTIPVGTAE
MUL_3190|M.ulcerans_Agy99 ---MPVPSSAVFECLLSLAAIKDVDARPTRTIDEVFTGKPLTTIPVGTAE
MSMEG_5912|M.smegmatis_MC2_155 ---MPAPSAADFARLRSLVAIEDLDARQSRPIEEVFTGRELTTIPVGTAE
MAV_4946|M.avium_104 MSDIQTQAKTEYDKLFIGGKWTEPSTSEVIEVHCPATGEYVGKVPLAAAA
MLBr_02573|M.leprae_Br4923 ---------------------------MSIATINPATGETVKTFTPTTQD
.* .
Mflv_0132|M.gilvum_PYR-GCK DVAQVAAGLRTAQRE--WEARGPKGRAKVLARYALWLGEHRAEIERLLIA
Mvan_0771|M.vanbaalenii_PYR-1 DVARLAAGLRTAQRE--WETRGPKGRAKVLARYAVWLGEHRDEIERLLIA
TH_3138|M.thermoresistible__bu DVPRITAGLREAQRH--WEARGPKGRAKVLARYAVWLGEHRDEIERLLIA
MAB_4136c|M.abscessus_ATCC_199 DIPGIVAQLAKAQPS--WERLGPEGRGQVLARFAQWLIDNTGRIEAQLIA
Rv1731|M.tuberculosis_H37Rv DVEAAFAEARAAQTD--WAKRPVIERAAVIRRYRDLVIENREFLMDLLQA
Mb1760|M.bovis_AF2122/97 DVEAAFAEARAAQTD--WAKRPVIERAAVIRRYRDLVIENREFLMDLLQA
MMAR_2571|M.marinum_M DVEAAFAEARAAQAN--WAKRPVSERAEVIRRYRDLVIENREFLMDLLQA
MUL_3190|M.ulcerans_Agy99 DVEAAFAEAREAQAN--WAKRPVSERAEVIRRYRDLVIENREFLMDLLQA
MSMEG_5912|M.smegmatis_MC2_155 DVAAAFAKARAAQRG--WAHRPVAERAAIMERFRDLVAKNRDFLMDVAQA
MAV_4946|M.avium_104 DVDAAVAAARAAFDNGPWPTTPPKERAAVIANALKVMEERKDLFTGLLAA
MLBr_02573|M.leprae_Br4923 EVEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQSAALMTL
:: * : *. : .
Mflv_0132|M.gilvum_PYR-GCK ETGKS-AVDAAQEVPLILMITSYYVRTMEKALAPESRPAALPFLSIKKIT
Mvan_0771|M.vanbaalenii_PYR-1 ETGKS-AADAAQEVPLIIMITSYYVRTMAKALAPETRPASLPFLSIKKIT
TH_3138|M.thermoresistible__bu ETGKS-AVDAAQEVPLILMITSYYIKTMEKALAPETRPASMPLLKIKKIT
MAB_4136c|M.abscessus_ATCC_199 ETGKS-KIDGGIEIPSILAIIAYYAPKAAEFLAPESRPASSIAMNTKKIT
Rv1731|M.tuberculosis_H37Rv EAGKA-RWAAQEEIVDLIANANYYARVCVDLLKP--RKAQPLLPGIGKTT
Mb1760|M.bovis_AF2122/97 EAGKA-RWAAQEEIVDLIANANYYARVCVDLLKP--RKAQPLLPGIGKTT
MMAR_2571|M.marinum_M EAGKA-RWAAQEEVVDLVANANYYARVSAGLLKP--RTVQALLPGIGKTT
MUL_3190|M.ulcerans_Agy99 EAGKA-RWAAQEEVVDLVANANYYARVSAGLLKP--RTVQALLPGIGKTT
MSMEG_5912|M.smegmatis_MC2_155 ETGKA-RSAAQEEIVDMMLNARYYARQAVKLLAP--KRVQGLLPGVVKTV
MAV_4946|M.avium_104 ETGQPPTSVETMHWMSSIGALNFFAGPAVDQVKWKEVRTG----GYGQSI
MLBr_02573|M.leprae_Br4923 EMGKT-LQSAKDEAMKCAKGFRYYAEKAETLLAD--EPAEAAAVGASKAL
* *:. . :: : . :
Mflv_0132|M.gilvum_PYR-GCK VHYRPRPVVGIIAPWNYPVANALMDAIGALAAGCAVLLKPSERTPLTAEL
Mvan_0771|M.vanbaalenii_PYR-1 VHYRPRPVVGIIAPWNYPVANALMDAVGALAAGCAVLLKPSERTPLTAEL
TH_3138|M.thermoresistible__bu VHHRPRPVVGVIAPWNYPVANALMDAIGALAAGCAVLLKPSERTPLTAEL
MAB_4136c|M.abscessus_ATCC_199 VHQRPRKVVGVISPWNYPVALGYWDVIPALLAGCSVLLKPSERTPLSDEL
Rv1731|M.tuberculosis_H37Rv VCYQPKGVVGVISPWNYPMTLTVSDSVPALVAGNAVVLKPDSQTPYCALA
Mb1760|M.bovis_AF2122/97 VCYQPKGVVGVISPWNYPMTLTVSDSVPALVAGNAVVLKPDSQTPYCALA
MMAR_2571|M.marinum_M VGYQPKGVVGVISPWNYPMTLTASDSVPALIAGNAVVLKPDSQTPYCALA
MUL_3190|M.ulcerans_Agy99 VGYQPKGVVGVISPWNYPMTLTASDSVPALIAGNAVVLKPDSQTPYCALA
MSMEG_5912|M.smegmatis_MC2_155 VNHHPKGVVGVISPWNYPMALSISDSIPALLAGNAVVVKPDSQTPYCTLA
MAV_4946|M.avium_104 VHREPIGVVGAIVAWNVPLFLAVNKLGPALLAGCTVVLKPAAETPLSANA
MLBr_02573|M.leprae_Br4923 ARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQCALY
. .* ** : .** *: ** ** ::* ..*
Mflv_0132|M.gilvum_PYR-GCK LLRGWLEAGGP-DVLALAQG-ARAVSEAVID--NSDFIQFTGSSATGAKV
Mvan_0771|M.vanbaalenii_PYR-1 LLRGWLDSGGP-DVLALAQG-AREVSEAVID--NSDYIQFTGSTATGVKV
TH_3138|M.thermoresistible__bu LLRGWLESGAP-EVFALAQG-AREVSEAVVD--NADFIQFTGSTATGIKV
MAB_4136c|M.abscessus_ATCC_199 IANGWAEIGGP-PVFEVVHG-AREVGEALVD--NVDFVQFTGSTATGKKI
Rv1731|M.tuberculosis_H37Rv CAELLYRAGLPRALYAIVPGPGSVVGTAITD--NCDYLMFTGSSATGSRL
Mb1760|M.bovis_AF2122/97 CAELLYRAGLPRALYAIVPGPGSVVGTAITD--NCDYLMFTGSSATGSRL
MMAR_2571|M.marinum_M CAELLYRAGLPRALYAIVPGPGSVVGTAIAD--NCDYLMFTGSSATGSRL
MUL_3190|M.ulcerans_Agy99 CAELLYRAGLPRALYAIVPGPGSVVGTAIAD--NCDYLMFTGSSATGSRL
MSMEG_5912|M.smegmatis_MC2_155 NAELLYEAGLPRDLFAVVPGPGSVVGTAIVE--NCDYLMFTGSTATGRTL
MAV_4946|M.avium_104 LAEAFAEAGLPEGVLSVVPG-GIETGQALTSNTDVDIFSFTGSSAVGREI
MLBr_02573|M.leprae_Br4923 LADVIARGGFPEDCFQTLLISANGVEAILRD-PRVAAATLTGSEPAGQAV
* * . . : . :*** ..* :
Mflv_0132|M.gilvum_PYR-GCK MERAARRLTPVSLELGGKDPMIVLEDADVDLAAHAAVWGAMFNAGQTCVS
Mvan_0771|M.vanbaalenii_PYR-1 MERAARRLTPVSLELGGKDPMIVLEDADIDLAAHAAVWGAMFNAGQTCVS
TH_3138|M.thermoresistible__bu MERAARRLTPVSLELGGKDPMIVLEDADIELAANAAVWGAMFNAGQTCVS
MAB_4136c|M.abscessus_ATCC_199 MERAARRLTPVSLELGGKDPMLVLPDADVKRAAHAAVWGSMFNAGQTCVS
Rv1731|M.tuberculosis_H37Rv AEHAGRRLIGFSAELGGKNPMIVARGANLDKVAKAATRACFSNAGQLCIS
Mb1760|M.bovis_AF2122/97 AEHAGRRLIGFSAELGGKNPMIVARGANLDKVAKAATRACFSNAGQLCIS
MMAR_2571|M.marinum_M AEHAGRRLIGFSAELGGKNAMIVTRGVNLDKVAKAATRACFSNAGQLCIS
MUL_3190|M.ulcerans_Agy99 AERAGRRLIGFSAELGGKNAMIVTRGVNLDKVAKAATRACFSNAGQLCIS
MSMEG_5912|M.smegmatis_MC2_155 AEQCGRRLIGFSAELGGKNPMIVTRGAKLDVAAKAATRACFSNAGQLCIS
MAV_4946|M.avium_104 GRRAADMLKPCTLELGGKSAAIVLDDVDLPSALPMLVFSGIMNSGQACVA
MLBr_02573|M.leprae_Br4923 GAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGRVQNNGQSCIA
.. : ****... :* ...: . . . . * ** *::
Mflv_0132|M.gilvum_PYR-GCK VERVYVLDQVYDQFVDAVVRDVAKLQVGAGEGN--AFGALIDDDQVAVTE
Mvan_0771|M.vanbaalenii_PYR-1 VERVYVLEQVYDQFVAAVVRDVESLKMGAGEGN--AFGALIDDSQVEITA
TH_3138|M.thermoresistible__bu VERVYVLAPVYEQFVAAVVRAVEKLEVGAGEGR--HFGALIDESQLAVTE
MAB_4136c|M.abscessus_ATCC_199 VERVYVHDSIYEQFVDLVVEEVRGLEIGAGLDH--SVGAMIDENQLEVVD
Rv1731|M.tuberculosis_H37Rv IERIYVEKDIAEEFTRKFGDAVRNMKLGTAYDFSVDMGSLISEAQLKTVS
Mb1760|M.bovis_AF2122/97 IERIYVEKDIAEEFTRKFGDAVRNMKLGTAYDFSVDMGSLISEAQLKTVS
MMAR_2571|M.marinum_M IERIYVEKDIADEFTRKFGDAVRSMKLGTAYDFSVDMGSLISEGQLKTVS
MUL_3190|M.ulcerans_Agy99 IERIYVEKDIADEFTRKFGDAVRSMKLGTAYDFSVDMGSLISEGQLKTVS
MSMEG_5912|M.smegmatis_MC2_155 IERIYVERAVADEFTAKFGEQVRSMRLAATYDFTADMGSLISEDQIKTVS
MAV_4946|M.avium_104 QTRILAPRSRYDEIVDAVSTFVQSLPVGLPSDPAAQIGSLISEKQRARVE
MLBr_02573|M.leprae_Br4923 AKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATESGRAQVE
*. . :::. . : : :. . .*.: : .
Mflv_0132|M.gilvum_PYR-GCK RHVADALAKGARALTGGTRGAGSG--SFYEPTVLVDVDHAMLCMTEETFG
Mvan_0771|M.vanbaalenii_PYR-1 RHVEDALAKGARALTGGKRGSGPG--SFYEPTVLVDVDHSMACMTEETFG
TH_3138|M.thermoresistible__bu RHVADAVARGAKVLTGGRRKDGPG--CFYEPTVLVDVDHSMMCMTEETFG
MAB_4136c|M.abscessus_ATCC_199 RHVRQAVSSGARALTGGARIPGHG--SFYAPTVLVDVDHSMDCMREETFG
Rv1731|M.tuberculosis_H37Rv GHVDDATAKGAKVIAGGKARPDIGP-LFYEPTVLTNVAPEMECAANETFG
Mb1760|M.bovis_AF2122/97 GHVDDATAKGAKVIAGGKARPDIGP-LFYEPTVLTNVAPEMECAANETFG
MMAR_2571|M.marinum_M GHVDDATAKGAKVIAGGKARPDVGP-LFYEPTVLTDVTHEMECADNETFG
MUL_3190|M.ulcerans_Agy99 GHVDDATAKGAKVIAGGKARPDVGP-LFYEPTVLTDVTHEMECADNETFG
MSMEG_5912|M.smegmatis_MC2_155 GHVDDAKAKGATVIAGGNIRPDIGP-RFYEPTVLTGVTDEMECARNETFG
MAV_4946|M.avium_104 GYIAKGIEEGARLVCGGGRPEGLDSGFFVQPTVFADVDNKMTIAQEEIFG
MLBr_02573|M.leprae_Br4923 QQVDAAAAAGAVIRCGGKRPDRPG--WFYPPTVITDITKDMALYTDEVFG
: . ** ** . * ***:..: * :* **
Mflv_0132|M.gilvum_PYR-GCK PTLPIMRVSTVDEAVRLANDSPYGLSAAVFSKDLERAEAVAVQLDCGGVN
Mvan_0771|M.vanbaalenii_PYR-1 PTLPIMKVSSVEEAVRLANDSPYGLSAAVFSRDVDRAKAVALQLDCGGVN
TH_3138|M.thermoresistible__bu PTLPIMKVSSVAEAVRLANDTPYGLSGSVFSQDVERAREIALQLDCGAVN
MAB_4136c|M.abscessus_ATCC_199 PTLPIMRYSEESEAIALANDSEYGLSASVWSKDRRRAERVALQLDCGTVN
Rv1731|M.tuberculosis_H37Rv PVVSIYPVADVDEAVEKANDTDYGLNASVWAGSTAEGQRIAARLRSGTVN
Mb1760|M.bovis_AF2122/97 PVVSIYPVADVDEAVEKANDTDYGLNASVWAGSTAEGQRIAARLRSGTVN
MMAR_2571|M.marinum_M PLVSIYPVADVEEAVEKANDTEYGLNASVWAGSTAEGEKIAARLRSGTVN
MUL_3190|M.ulcerans_Agy99 PLVSIYPVADVEEAVEKANDTEYGLNASVWAGSTPEGEKIAARLRSGTVN
MSMEG_5912|M.smegmatis_MC2_155 PVVSIYPVESVAEAIEKANDTEYGLNASVWAGSKTEGEAIAAQLQAGTVN
MAV_4946|M.avium_104 PVLSIIPYDTEEDAIKIANDSVYGLAGSVWTADVQRGIAVSEKIRTGTYA
MLBr_02573|M.leprae_Br4923 PVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNIVAGQVF
* .: :*: ** : :** . .:: . . . .: *
Mflv_0132|M.gilvum_PYR-GCK IND-VISNLMCTTAPMGGWKTSGIGARFGGPEGLRKYCRIETVVAPRTNV
Mvan_0771|M.vanbaalenii_PYR-1 VND-VISNLMCTTAPMGGWKSSGIGARFGGPEGLRKYCRIETVVSPRTNV
TH_3138|M.thermoresistible__bu IND-VISNLMATTAPMGGWKTSGIGARFGGPEGLRKYCRVETVVEPRTGI
MAB_4136c|M.abscessus_ATCC_199 IND-VIMNLSCLTAPHGGWKGSGLGSRFGGVDGLRKYCRTEAIVDTRVTL
Rv1731|M.tuberculosis_H37Rv VDEGYAFAWGSLSAPMGGMGLSGVGRRHG-PEGLLKYTESQTIATARVFN
Mb1760|M.bovis_AF2122/97 VDEGYAFAWGSLSAPMGGMGLSGVGRRHG-PEGLLKYTESQTIATARVFN
MMAR_2571|M.marinum_M VNEGYAFAWGSLSAPMGGMGISGVGRRHG-PEGLLKYTESQTIATARVFN
MUL_3190|M.ulcerans_Agy99 VNEGYAFAWGSLSAPMGGMGISGVGRRHG-PEGLLKYTESQTIATARVFN
MSMEG_5912|M.smegmatis_MC2_155 VDEGYALAFGSTAAPMGGMKASGVGRRHG-ADGILKYTESQTVATSRVLN
MAV_4946|M.avium_104 INW----YAFDPCCPFGGYKNSGIGRENG-PEGVEHFTQQKSVLMPMGYT
MLBr_02573|M.leprae_Br4923 ING---MTVSYPELPFGGIKRSGYGRELS-THGIREFCNIKTVWIA----
:: * ** ** * . . .*: .: . ::: .
Mflv_0132|M.gilvum_PYR-GCK GAGGNYYNNSPRALKRMNTMMTKLALIRPRRVAK
Mvan_0771|M.vanbaalenii_PYR-1 GAGGNYYNNSLRALKRMNKLMTKLALIRPRRVAK
TH_3138|M.thermoresistible__bu GAGGTYYNNSFKSLGMMNKLLTRMALVRPRRLAK
MAB_4136c|M.abscessus_ATCC_199 PSEPLWYSGPSWLAPVALKSLTKLSNKALRPFRR
Rv1731|M.tuberculosis_H37Rv LDPPFGIPATVWQKSLLPIVRTVMKLPGRR----
Mb1760|M.bovis_AF2122/97 LDPPFGIPATVWQKSLLPIVRTVMKLPGRR----
MMAR_2571|M.marinum_M LDPPMGVPPTLWQKSLLPIVRTVMKLPGRK----
MUL_3190|M.ulcerans_Agy99 LDPPMGVPPTLWQKSLLPIVRTVMKLPGRK----
MSMEG_5912|M.smegmatis_MC2_155 LDPPLGISGTLWQKAMTPMIRAVQKLPGR-----
MAV_4946|M.avium_104 LEG-------------------------------
MLBr_02573|M.leprae_Br4923 ----------------------------------