For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTTSENLVSTPSDTSHDVRNPANGQVVGRVNWTDPSDIPGIVAQLAKAQPSWERLGPEGRGQVLARFAQW LIDNTGRIEAQLIAETGKSKIDGGIEIPSILAIIAYYAPKAAEFLAPESRPASSIAMNTKKITVHQRPRK VVGVISPWNYPVALGYWDVIPALLAGCSVLLKPSERTPLSDELIANGWAEIGGPPVFEVVHGAREVGEAL VDNVDFVQFTGSTATGKKIMERAARRLTPVSLELGGKDPMLVLPDADVKRAAHAAVWGSMFNAGQTCVSV ERVYVHDSIYEQFVDLVVEEVRGLEIGAGLDHSVGAMIDENQLEVVDRHVRQAVSSGARALTGGARIPGH GSFYAPTVLVDVDHSMDCMREETFGPTLPIMRYSEESEAIALANDSEYGLSASVWSKDRRRAERVALQLD CGTVNINDVIMNLSCLTAPHGGWKGSGLGSRFGGVDGLRKYCRTEAIVDTRVTLPSEPLWYSGPSWLAPV ALKSLTKLSNKALRPFRR*
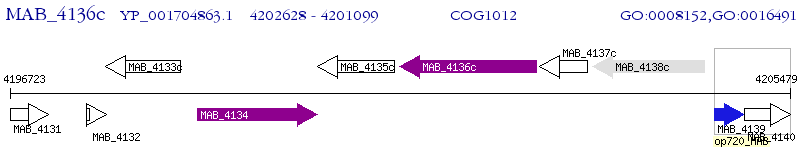
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4136c | - | - | 100% (509) | putative aldehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4484 | - | 2e-71 | 37.90% (467) | aldehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 1e-70 | 36.75% (449) | gamma-aminobutyraldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1760 | gabD2 | 6e-79 | 37.89% (475) | succinic semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0132 | - | 1e-174 | 60.39% (510) | aldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv1731 | gabD2 | 6e-79 | 37.89% (475) | succinic semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 2e-46 | 29.14% (453) | succinic semialdehyde dehydrogenase |
| M. marinum M | MMAR_3111 | gabD1_1 | 1e-122 | 48.25% (487) | succinate-semialdehyde dehydrogenase |
| M. avium 104 | MAV_4946 | - | 2e-68 | 36.29% (463) | aldehyde dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_0889 | - | 1e-170 | 58.40% (500) | succinic semialdehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_3138 | - | 1e-170 | 61.63% (490) | PUTATIVE aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_2361 | gabD1_1 | 1e-121 | 48.25% (487) | succinate-semialdehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0771 | - | 1e-171 | 59.41% (510) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1760|M.bovis_AF2122/97 -------------MPAPSAEVFDRLRNLAAIKDVAARPTRTIDEVFTGKP
Rv1731|M.tuberculosis_H37Rv -------------MPAPSAEVFDRLRNLAAIKDVAARPTRTIDEVFTGKP
Mflv_0132|M.gilvum_PYR-GCK ----------------------MTATSDAHTANPFTGTGTITN-PATGAV
Mvan_0771|M.vanbaalenii_PYR-1 -------------------MGFMTATSDVQPPHTFTGTGTIRN-PATGAV
MSMEG_0889|M.smegmatis_MC2_155 ----------------------MTATPEVDLPNPYTGTGTIHN-PATGAV
TH_3138|M.thermoresistible__bu ----------------------MTATSGVDA----TETATIRN-PATGGV
MAB_4136c|M.abscessus_ATCC_199 ----------------------MTTTSENLVSTPSDTSHDVRN-PANGQV
MMAR_3111|M.marinum_M MVCWVPYEQLRLAPAAKAVAVMMDTTNQANPAEADTRDEVVVKCPATGEI
MUL_2361|M.ulcerans_Agy99 MVCWVPYEQLRLAPAAKAVAVMMDTTNQANPAEADTRDEVVVKCPATGEI
MAV_4946|M.avium_104 ----------MSDIQTQAKTEYDKLFIGGKWTEPSTSEVIEVHCPATGEY
MLBr_02573|M.leprae_Br4923 -------------------------------------MSIATINPATGET
.*
Mb1760|M.bovis_AF2122/97 LTTIPVGTAADVEAAFAEARAAQT--DWAKRPVIERAAVIRRYRDLVIEN
Rv1731|M.tuberculosis_H37Rv LTTIPVGTAADVEAAFAEARAAQT--DWAKRPVIERAAVIRRYRDLVIEN
Mflv_0132|M.gilvum_PYR-GCK AGTVHWTDPADVAQVAAGLRTAQR--EWEARGPKGRAKVLARYALWLGEH
Mvan_0771|M.vanbaalenii_PYR-1 AGEVRWTDPADVARLAAGLRTAQR--EWETRGPKGRAKVLARYAVWLGEH
MSMEG_0889|M.smegmatis_MC2_155 AGEVRWTDPADVPRIAAGLREAQK--EWEARGPKGRAKVLARYAVWLGDH
TH_3138|M.thermoresistible__bu AGEVRWTDPADVPRITAGLREAQR--HWEARGPKGRAKVLARYAVWLGEH
MAB_4136c|M.abscessus_ATCC_199 VGRVNWTDPSDIPGIVAQLAKAQP--SWERLGPEGRGQVLARFAQWLIDN
MMAR_3111|M.marinum_M VGTVAVTTVDGVAAAAARLRAAQP--EWQRLGVSGRAYWLGRWRDWMLDH
MUL_2361|M.ulcerans_Agy99 VGTVPVTTVDGVAAAAARLRAAQP--EWQRLGVSGRACWLGRWRDWMLDH
MAV_4946|M.avium_104 VGKVPLAAAADVDAAVAAARAAFDNGPWPTTPPKERAAVIANALKVMEER
MLBr_02573|M.leprae_Br4923 VKTFTPTTQDEVEDAIARAYARFD--NYRHTTFTQRAKWANATADLLEAE
. : * : *. : .
Mb1760|M.bovis_AF2122/97 REFLMDLLQAEAGKA-RWAAQEEIVDLIANANYYARVCVDLLKP--RKAQ
Rv1731|M.tuberculosis_H37Rv REFLMDLLQAEAGKA-RWAAQEEIVDLIANANYYARVCVDLLKP--RKAQ
Mflv_0132|M.gilvum_PYR-GCK RAEIERLLIAETGKS-AVDAAQEVPLILMITSYYVRTMEKALAPESRPAA
Mvan_0771|M.vanbaalenii_PYR-1 RDEIERLLIAETGKS-AADAAQEVPLIIMITSYYVRTMAKALAPETRPAS
MSMEG_0889|M.smegmatis_MC2_155 RDEIEKLLIAETGKS-AVDAAQEVPLILMILSYYIKTVEKAMAPEKRPAP
TH_3138|M.thermoresistible__bu RDEIERLLIAETGKS-AVDAAQEVPLILMITSYYIKTMEKALAPETRPAS
MAB_4136c|M.abscessus_ATCC_199 TGRIEAQLIAETGKS-KIDGGIEIPSILAIIAYYAPKAAEFLAPESRPAS
MMAR_3111|M.marinum_M RDDLLTPLQLETGKS-WGDAGIEIATVQVIN-YWIDNAAEFLADQPVRPY
MUL_2361|M.ulcerans_Agy99 RDDLLTPLQLETGKS-WGDAGIEIATVQVIN-YWIDNAAEFLADQPVRPY
MAV_4946|M.avium_104 KDLFTGLLAAETGQPPTSVETMHWMSSIGALNFFAGPAVDQVKWKEVRTG
MLBr_02573|M.leprae_Br4923 ANQSAALMTLEMGKT-LQSAKDEAMKCAKGFRYYAEKAETLLAD--EPAE
: * *:. . :: : .
Mb1760|M.bovis_AF2122/97 PLLPGIGKTTVCYQPKGVVGVISPWNYPMTLTVSDSVPALVAGNAVVLKP
Rv1731|M.tuberculosis_H37Rv PLLPGIGKTTVCYQPKGVVGVISPWNYPMTLTVSDSVPALVAGNAVVLKP
Mflv_0132|M.gilvum_PYR-GCK LPFLSIKKITVHYRPRPVVGIIAPWNYPVANALMDAIGALAAGCAVLLKP
Mvan_0771|M.vanbaalenii_PYR-1 LPFLSIKKITVHYRPRPVVGIIAPWNYPVANALMDAVGALAAGCAVLLKP
MSMEG_0889|M.smegmatis_MC2_155 LPFLSIKKIEVHYRPRAVVGIIAPWNYPVANAMMDAIGALAAGCSVLLKP
TH_3138|M.thermoresistible__bu MPLLKIKKITVHHRPRPVVGVIAPWNYPVANALMDAIGALAAGCAVLLKP
MAB_4136c|M.abscessus_ATCC_199 SIAMNTKKITVHQRPRKVVGVISPWNYPVALGYWDVIPALLAGCSVLLKP
MMAR_3111|M.marinum_M GAANAAKKLTIVYEPYPLVGVITPWNGPLSVPLLDVPAALLAGCAVLSKP
MUL_2361|M.ulcerans_Agy99 GAANAAKKLTIVYEPYPLVGVITPWNGPLSVPLLDVPAALLAGCAVLSKP
MAV_4946|M.avium_104 ----GYGQSIVHREPIGVVGAIVAWNVPLFLAVNKLGPALLAGCTVVLKP
MLBr_02573|M.leprae_Br4923 AAAVGASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKH
: .* :* : .** *: ** ** : *
Mb1760|M.bovis_AF2122/97 DSQTPYCALACAELLYRAGLPRALYAIVPGPGSVVGTAITD--NCDYLMF
Rv1731|M.tuberculosis_H37Rv DSQTPYCALACAELLYRAGLPRALYAIVPGPGSVVGTAITD--NCDYLMF
Mflv_0132|M.gilvum_PYR-GCK SERTPLTAELLLRGWLEAGGP-DVLALAQGAR-AVSEAVID--NSDFIQF
Mvan_0771|M.vanbaalenii_PYR-1 SERTPLTAELLLRGWLDSGGP-DVLALAQGAR-EVSEAVID--NSDYIQF
MSMEG_0889|M.smegmatis_MC2_155 SERTPLTAEVLLRGWLDSGAP-EVFALAQGAR-AVSEAVID--NSDYIQF
TH_3138|M.thermoresistible__bu SERTPLTAELLLRGWLESGAP-EVFALAQGAR-EVSEAVVD--NADFIQF
MAB_4136c|M.abscessus_ATCC_199 SERTPLSDELIANGWAEIGGP-PVFEVVHGAR-EVGEALVD--NVDFVQF
MMAR_3111|M.marinum_M SEFTPLAWQHAVEGWKQIGAP-DVLDVVFGFG-ETGAALVD--VVDYVMF
MUL_2361|M.ulcerans_Agy99 SEFTPLAWQHAVEGWKQIGAP-DVLDVVFGFG-ETGAALVD--VVDYVMF
MAV_4946|M.avium_104 AAETPLSANALAEAFAEAGLPEGVLSVVPGGI-ETGQALTSNTDVDIFSF
MLBr_02573|M.leprae_Br4923 ASNVPQCALYLADVIARGGFPEDCFQTLLISANGVEAILRD-PRVAAATL
.* * * . : . :
Mb1760|M.bovis_AF2122/97 TGSSATGSRLAEHAGRRLIGFSAELGGKNPMIVARGANLDKVAKAATRAC
Rv1731|M.tuberculosis_H37Rv TGSSATGSRLAEHAGRRLIGFSAELGGKNPMIVARGANLDKVAKAATRAC
Mflv_0132|M.gilvum_PYR-GCK TGSSATGAKVMERAARRLTPVSLELGGKDPMIVLEDADVDLAAHAAVWGA
Mvan_0771|M.vanbaalenii_PYR-1 TGSTATGVKVMERAARRLTPVSLELGGKDPMIVLEDADIDLAAHAAVWGA
MSMEG_0889|M.smegmatis_MC2_155 TGSSATGAKVMERAARRLTPVSLELGGKDPMIVLEDADIELAANAAVWGA
TH_3138|M.thermoresistible__bu TGSTATGIKVMERAARRLTPVSLELGGKDPMIVLEDADIELAANAAVWGA
MAB_4136c|M.abscessus_ATCC_199 TGSTATGKKIMERAARRLTPVSLELGGKDPMLVLPDADVKRAAHAAVWGS
MMAR_3111|M.marinum_M TGSVGTGRRIAVAAAERLIPCGLELGGKDAMIVCADADIDRAVDGAVWGG
MUL_2361|M.ulcerans_Agy99 TGSVGTGRRIAVAAAERLIPCGLELGGKDAMIVCADADIDRAVDGAVWGG
MAV_4946|M.avium_104 TGSSAVGREIGRRAADMLKPCTLELGGKSAAIVLDDVDLPSALPMLVFSG
MLBr_02573|M.leprae_Br4923 TGSEPAGQAVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAVATAVTGR
*** .* : *. : ****... :* ..:: . . .
Mb1760|M.bovis_AF2122/97 FSNAGQLCISIERIYVEKDIAEEFTRKFGDAVRNMKLGTAYDFS--VDMG
Rv1731|M.tuberculosis_H37Rv FSNAGQLCISIERIYVEKDIAEEFTRKFGDAVRNMKLGTAYDFS--VDMG
Mflv_0132|M.gilvum_PYR-GCK MFNAGQTCVSVERVYVLDQVYDQFVDAVVRDVAKLQVGAGEGN----AFG
Mvan_0771|M.vanbaalenii_PYR-1 MFNAGQTCVSVERVYVLEQVYDQFVAAVVRDVESLKMGAGEGN----AFG
MSMEG_0889|M.smegmatis_MC2_155 MFNAGQTCVSVERVYVLESVYDQFVAATVRAVENLKMGAGEGY----DFG
TH_3138|M.thermoresistible__bu MFNAGQTCVSVERVYVLAPVYEQFVAAVVRAVEKLEVGAGEGR----HFG
MAB_4136c|M.abscessus_ATCC_199 MFNAGQTCVSVERVYVHDSIYEQFVDLVVEEVRGLEIGAGLDH----SVG
MMAR_3111|M.marinum_M FAYTGQVCISVERIYVEAPVFDTFVSKLVARTSQLRQGMDAGHDYSCEIG
MUL_2361|M.ulcerans_Agy99 FAYTGQVCISVERIYVEALVFDTFVSKLVARTAQLRQGMDAGRDYSCEIG
MAV_4946|M.avium_104 IMNSGQACVAQTRILAPRSRYDEIVDAVSTFVQSLPVGLPSDP--AAQIG
MLBr_02573|M.leprae_Br4923 VQNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPD--TDVG
. ** *:: *. . : :. : * . .*
Mb1760|M.bovis_AF2122/97 SLISEAQLKTVSGHVDDATAKGAKVIAGGKARP-DIGPLFYEPTVLTNVA
Rv1731|M.tuberculosis_H37Rv SLISEAQLKTVSGHVDDATAKGAKVIAGGKARP-DIGPLFYEPTVLTNVA
Mflv_0132|M.gilvum_PYR-GCK ALIDDDQVAVTERHVADALAKGARALTGGTRGA--GSGSFYEPTVLVDVD
Mvan_0771|M.vanbaalenii_PYR-1 ALIDDSQVEITARHVEDALAKGARALTGGKRGS--GPGSFYEPTVLVDVD
MSMEG_0889|M.smegmatis_MC2_155 SLIDAGQVDITERHVNEALEKGAKALTGGKRPE--DGGSFYPPTVLVDVD
TH_3138|M.thermoresistible__bu ALIDESQLAVTERHVADAVARGAKVLTGGRRKD--GPGCFYEPTVLVDVD
MAB_4136c|M.abscessus_ATCC_199 AMIDENQLEVVDRHVRQAVSSGARALTGGARIP--GHGSFYAPTVLVDVD
MMAR_3111|M.marinum_M SMTTAQQLEIVSRHVDDAVRKGAKVLIGGKTRDPGGDGLFYEPTVLVDVD
MUL_2361|M.ulcerans_Agy99 SMTAAQQLEIVSRHVDDAVCKGAKVLIGGKTRDPGGDGLFYKPTVLVDVD
MAV_4946|M.avium_104 SLISEKQRARVEGYIAKGIEEGARLVCGGGRPEGLDSGFFVQPTVFADVD
MLBr_02573|M.leprae_Br4923 PLATESGRAQVEQQVDAAAAAGAVIRCGGKRPD--RPGWFYPPTVITDIT
.: . : . ** ** * ***:.::
Mb1760|M.bovis_AF2122/97 PEMECAANETFGPVVSIYPVADVDEAVEKANDTDYGLNASVWAGSTAEGQ
Rv1731|M.tuberculosis_H37Rv PEMECAANETFGPVVSIYPVADVDEAVEKANDTDYGLNASVWAGSTAEGQ
Mflv_0132|M.gilvum_PYR-GCK HAMLCMTEETFGPTLPIMRVSTVDEAVRLANDSPYGLSAAVFSKDLERAE
Mvan_0771|M.vanbaalenii_PYR-1 HSMACMTEETFGPTLPIMKVSSVEEAVRLANDSPYGLSAAVFSRDVDRAK
MSMEG_0889|M.smegmatis_MC2_155 HSMSCMNEETFGPTLPIMKVSSVEEAVRLANDSPYGLSASVFSKDTERAK
TH_3138|M.thermoresistible__bu HSMMCMTEETFGPTLPIMKVSSVAEAVRLANDTPYGLSGSVFSQDVERAR
MAB_4136c|M.abscessus_ATCC_199 HSMDCMREETFGPTLPIMRYSEESEAIALANDSEYGLSASVWSKDRRRAE
MMAR_3111|M.marinum_M HDMDCMREETFGPTLPVMKVRDVAEAIEKANDCRFGLSGSVWTRDKAKAI
MUL_2361|M.ulcerans_Agy99 HDMDCMREETFGPTLPVMKVRDVAEAIEKANDCRFGLSGSVWTRDKAKAI
MAV_4946|M.avium_104 NKMTIAQEEIFGPVLSIIPYDTEEDAIKIANDSVYGLAGSVWTADVQRGI
MLBr_02573|M.leprae_Br4923 KDMALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQR
* :* ***. .: :*: ** :** . .:: . .
Mb1760|M.bovis_AF2122/97 RIAARLRSGTVNVDEGYAFAWGSLSAPMGGMGLSGVGRRHG-PEGLLKYT
Rv1731|M.tuberculosis_H37Rv RIAARLRSGTVNVDEGYAFAWGSLSAPMGGMGLSGVGRRHG-PEGLLKYT
Mflv_0132|M.gilvum_PYR-GCK AVAVQLDCGGVNIND-VISNLMCTTAPMGGWKTSGIGARFGGPEGLRKYC
Mvan_0771|M.vanbaalenii_PYR-1 AVALQLDCGGVNVND-VISNLMCTTAPMGGWKSSGIGARFGGPEGLRKYC
MSMEG_0889|M.smegmatis_MC2_155 KIALQLDCGAVNIND-VISNLMCTTAPMGGWKTSGIGARFGGVDGVRKFC
TH_3138|M.thermoresistible__bu EIALQLDCGAVNIND-VISNLMATTAPMGGWKTSGIGARFGGPEGLRKYC
MAB_4136c|M.abscessus_ATCC_199 RVALQLDCGTVNIND-VIMNLSCLTAPHGGWKGSGLGSRFGGVDGLRKYC
MMAR_3111|M.marinum_M DIARRLNTGTVNINN-VITGIFQIGVPMAGWKESGLGTRSGGAEGIRKYC
MUL_2361|M.ulcerans_Agy99 DIARRLNTGTVNINN-VITGIFQIGVPMAGWKESGLGTRSGGAEGIRKYC
MAV_4946|M.avium_104 AVSEKIRTGTYAINW----YAFDPCCPFGGYKNSGIGRENG-PEGVEHFT
MLBr_02573|M.leprae_Br4923 HFIDNIVAGQVFING---MTVSYPELPFGGIKRSGYGRELS-THGIREFC
. .: * :: * .* ** * . . .*: .:
Mb1760|M.bovis_AF2122/97 ESQTIATARVFNLDPPFGIPATVWQKSLLPIVRTVMKLPGRR-----
Rv1731|M.tuberculosis_H37Rv ESQTIATARVFNLDPPFGIPATVWQKSLLPIVRTVMKLPGRR-----
Mflv_0132|M.gilvum_PYR-GCK RIETVVAPRTNVGAGGNYYNNSPRALKRMNTMMTKLALIRPRR-VAK
Mvan_0771|M.vanbaalenii_PYR-1 RIETVVSPRTNVGAGGNYYNNSLRALKRMNKLMTKLALIRPRR-VAK
MSMEG_0889|M.smegmatis_MC2_155 KQETVVAPRTSVGAGGTYYNNSHKALARMNRLMTKLALARPRR-TAK
TH_3138|M.thermoresistible__bu RVETVVEPRTGIGAGGTYYNNSFKSLGMMNKLLTRMALVRPRR-LAK
MAB_4136c|M.abscessus_ATCC_199 RTEAIVDTRVTLPSEPLWYSGPSWLAPVALKSLTKLSNKALRP-FRR
MMAR_3111|M.marinum_M HPKSIVAERVAMKKELLWYPYSRKKGSFAEKVVRMLSARDWRRRLGR
MUL_2361|M.ulcerans_Agy99 HPKSIVAERVAMKKELLWYPYSRKNGSFAEKVVKMLSARDWRRRLGR
MAV_4946|M.avium_104 QQKSVLMPMGYTLEG--------------------------------
MLBr_02573|M.leprae_Br4923 NIKTVWIA---------------------------------------
. :::