For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EGTIAVPGGRVWFQRIGGGPGRPLLVVHGGPGLPHNYLAPLRRLSDEREVIFWDQLGCGNSACPSDVDLW TMNRSVAEMATVAEALALTRFHIFSHSWGGMLAQQYVLDKAPDAVSLTIANSTASIPEFSASLVSLKSCL DVATRSAIDRHEAAGTTHSAEYQAAIRTWNETYLCRTRPWPRELTEAFANMGTEIFETMFGPSDFRIVGN VRDWDVVDRLADIAVPTLLVVGRFDECSPEHMREMQGRIAGSRLEFFESSSHMPFIEEPARFDRVMREFL RLHDI*
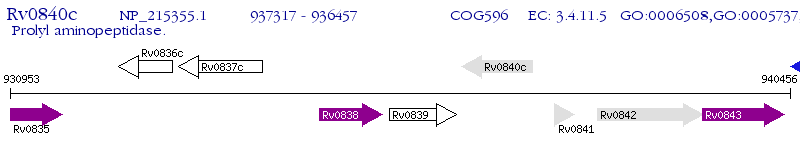
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0840c | pip | - | 100% (286) | PROBABLE PROLINE IMINOPEPTIDASE PIP (PROLYL AMINOPEPTIDASE) (PAP) |
| M. tuberculosis H37Rv | Rv2715 | - | 2e-07 | 26.17% (298) | hydrolase |
| M. tuberculosis H37Rv | Rv2296 | - | 5e-05 | 26.42% (106) | haloalkane dehalogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0863c | pip | 1e-169 | 100.00% (286) | proline iminopeptidase |
| M. gilvum PYR-GCK | Mflv_2400 | - | 3e-08 | 23.90% (272) | alpha/beta hydrolase fold |
| M. leprae Br4923 | MLBr_00862 | ephD | 1e-06 | 26.73% (101) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3934c | - | 2e-06 | 23.70% (270) | alpha/beta fold hydrolase |
| M. marinum M | MMAR_4789 | pip | 1e-125 | 75.00% (284) | proline iminopeptidase Pip |
| M. avium 104 | MAV_0868 | - | 1e-121 | 72.34% (282) | proline imino-peptidase |
| M. smegmatis MC2 155 | MSMEG_2681 | - | 3e-54 | 39.41% (269) | proline imino-peptidase |
| M. thermoresistible (build 8) | TH_4528 | - | 1e-08 | 22.57% (288) | PUTATIVE alpha/beta hydrolase fold |
| M. ulcerans Agy99 | MUL_0358 | pip | 1e-107 | 73.05% (256) | proline iminopeptidase PIP |
| M. vanbaalenii PYR-1 | Mvan_2912 | - | 2e-08 | 25.26% (285) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0840c|M.tuberculosis_H37Rv ----------------MEGTIAVP-GGRVWFQRIGGG----PGRPLLVVH
Mb0863c|M.bovis_AF2122/97 ----------------MEGTIAVP-GGRVWFQRIGGG----PGRPLLVVH
MMAR_4789|M.marinum_M -------------MTKPEATIAVP-GGKVWSKRVGGG----SGLPLLVVH
MUL_0358|M.ulcerans_Agy99 -------------MEQARRRRLRP-SPARGAWRPGPA----ALLP-----
MAV_0868|M.avium_104 -------------------MVAVP-GGQVWFQRTGGG----AGLPLLVIH
MSMEG_2681|M.smegmatis_MC2_155 ---------MLSRMPVSSRTVPFG-DHETWVQVTTPENAQPHALPLIVLH
Mflv_2400|M.gilvum_PYR-GCK MAEFESVWSDLQGVAFSQGYLDAG-GVRTRYLHAGDP----ADPALVFLH
TH_4528|M.thermoresistible__bu ------VWSDLQGVAFTQGYLDAG-GVRTRYLHAGEA----DKPVLIFLH
Mvan_2912|M.vanbaalenii_PYR-1 -------------MHDDVKYLDLH-GDRVAYREAGQG------PAVLLIH
MAB_3934c|M.abscessus_ATCC_199 -----------------MPLANIN-GIQISYDDRGPLALGASGDPVVFIS
MLBr_00862|M.leprae_Br4923 ------MPATQSIHKVPHQFVESPDGVRLAIYQSGQP----EGPAIVLVH
.
Rv0840c|M.tuberculosis_H37Rv GGPGLPHNYLAPLRRLSDE--REVIFWDQLGCGNSACPSDV--DLWTMNR
Mb0863c|M.bovis_AF2122/97 GGPGLPHNYLAPLRRLSDE--REVIFWDQLGCGNSACPSDV--DLWTMNR
MMAR_4789|M.marinum_M GGPGLPHYYLSALQRLAGE--REVIFWDQLGCGNSERPSDV--DLWTMGR
MUL_0358|M.ulcerans_Agy99 -------DYLSALQRLAGE--REVIFWDQLGCGNSERPSDV--DLWTMGR
MAV_0868|M.avium_104 GGPGLPHDYLRSLRRLATD--REVIFWDQLGCGNSKCPPNP--GLWTMER
MSMEG_2681|M.smegmatis_MC2_155 GGPGMAHNYVANIAALADETGRTVIHYDQVGCGNSTHLPDAPADFWTPQL
Mflv_2400|M.gilvum_PYR-GCK GSGGHAEAYVRNLEAHAEH--FSTWSIDMLGHGYTDKPGHP----LEIPH
TH_4528|M.thermoresistible__bu GSGGHAEAYVRNLEAHAEH--FSTWSIDMLGHGYTDKPGHP----LEVRH
Mvan_2912|M.vanbaalenii_PYR-1 GMGGSSLTWKALLPHLATR--YRVIAPDLLGHGQSDKPRGD----YSLGA
MAB_3934c|M.abscessus_ATCC_199 GRGGAGRSWHLHQVPAFRAAGYRTITFNNRGIPPTTESADG----FTLQD
MLBr_00862|M.leprae_Br4923 GFPDSHVLWDGVVPLLAKR--FRIVRYDNRGVGQSSVPKPVS--AYTMTR
: : * :
Rv0840c|M.tuberculosis_H37Rv SVAEMATVAEALALTR-FHIFSHSWGGMLAQQYVLDKAPD--AVSLTIAN
Mb0863c|M.bovis_AF2122/97 SVAEMATVAEALALTR-FHIFSHSWGGMLAQQYVLDKAPD--AVSLTIAN
MMAR_4789|M.marinum_M SVAEMDAVVRALGLEA-FHIFGNSWGGMLAQQYVLDVPSG--AVSLTISN
MUL_0358|M.ulcerans_Agy99 SVAEIDAVVRALGLEA-FHIFGNSWGGMLAQQYVLDVPSG--AVSLTISN
MAV_0868|M.avium_104 SVAEVDAVVRALRLDR-FHLFGNSWGGMLAQQYVLDAAPAG-AASLTISN
MSMEG_2681|M.smegmatis_MC2_155 FVDEFHAVCTALGIER-YHVLGQSWGGMLGAEIAVRQPSG--LVSLAICN
Mflv_2400|M.gilvum_PYR-GCK YVDHLAAFLDAIGARR-AHISGESLGGWVAARIAADRPER--VDRLVLNT
TH_4528|M.thermoresistible__bu YVEHLAAVLDAIGADR-AHISGESLGGWVASRFAIDRPDR--LDRLVLNT
Mvan_2912|M.vanbaalenii_PYR-1 FAVWLRDLLDLLGIAR-VTLVGHSLGGGVAMQFVHQHPDY--CERLVLIS
MAB_3934c|M.abscessus_ATCC_199 MVDDTAALIEQLGAAP-ARLVGFSMGALIAQELTLTRPEL--VTAAVFMG
MLBr_00862|M.leprae_Br4923 FADDFAAVIDELSPDRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVS
. . : : . . *. . .
Rv0840c|M.tuberculosis_H37Rv STASIPEFSASLVSLKSC-----LDVATRSAIDRHEAAGTTHSAEYQAAI
Mb0863c|M.bovis_AF2122/97 STASIPEFSASLVSLKSC-----LDVATRSAIDRHEAAGTTHSAEYQAAI
MMAR_4789|M.marinum_M STASIPRFADNVIRLKAE-----LDPGTQATIDRHEAAGTTHSAEYQSAI
MUL_0358|M.ulcerans_Agy99 STASIPRFADSVIRLKAE-----LDPGTQATIDRHEAAGTTHSAEYQSAI
MAV_0868|M.avium_104 SIASIPQFGKMVARLKSE-----LDPATQAAIDRHEAAGTTHDPEYQAAI
MSMEG_2681|M.smegmatis_MC2_155 SPASMRLWSEAAGDLRAQ-----LPAETRAALDRHEAAGTITHPDYLQAA
Mflv_2400|M.gilvum_PYR-GCK AGGSQADPEVMKRIITLS-----MAAAENPTWDTVQARIKWLMADKSKDY
TH_4528|M.thermoresistible__bu AGGSQADPEVMKRIITLS-----MAAAENPSWDTVQARITWLMADKSKGY
Mvan_2912|M.vanbaalenii_PYR-1 SGGLGPELGRSLRLLSTPGIELLLPVAAAPSVVAVGERVRSWLGARGLAS
MAB_3934c|M.abscessus_ATCC_199 TRGREDATRSFFRKAELE-----LSASGVEVPAAYQAAMRLLLNFSPKTL
MLBr_00862|M.leprae_Br4923 GPSQDQLVDYILSGLRRPWR--PRTFARAVSQLFRLTYMVLFSIPVLVPM
.
Rv0840c|M.tuberculosis_H37Rv RTWNET--YLCRTRPWPRELTEAFANMGTE--IFETMFGPSDFRIVGNVR
Mb0863c|M.bovis_AF2122/97 RTWNET--YLCRTRPWPRELTEAFANMGTE--IFETMFGPSDFRIVGNVR
MMAR_4789|M.marinum_M TTWNET--HLCRTLPWPAYLTEAFQNLGPE--IFETMFGPSDFRIVGTIR
MUL_0358|M.ulcerans_Agy99 TTWNET--YLCRTLPWPAYLTEAFQNLGPE--IFETMFGPSDFRIVGTIR
MAV_0868|M.avium_104 RTWNET--YLCRVRPWPRDLEDAFRKMSAE--VFETMFGASDFHIVGTVR
MSMEG_2681|M.smegmatis_MC2_155 AEFYRR--HVCRVVPTPQDFADSVAQMEAEPTVYHTMNGPNEFHVVGTLG
Mflv_2400|M.gilvum_PYR-GCK DDIVASRQRVYRQPGFANAMRDIMALQDPE-IRQRNLLGPNEY-------
TH_4528|M.thermoresistible__bu DDIVASRQKIYRQPGFVDAMRNIMALQDPE-IRARNLLGPEEY-------
Mvan_2912|M.vanbaalenii_PYR-1 PEVGET-WNAYASLSDPQTRTAFLRTLRSVVDAQGQAVSALNR-------
MAB_3934c|M.abscessus_ATCC_199 NNDAAIKDWIDMFTLWPEPASGGIDHQRSA---IPAPDRPGAY-------
MLBr_00862|M.leprae_Br4923 VLWIALSSATIRRTMVDNISTDQIHHSATLARDAAHSVKTYPANYFRAFS
. . .
Rv0840c|M.tuberculosis_H37Rv DWDVVDRLADIAVPTLLVVGRFD-ECSPEHMREMQGRIAGSRLEFFESSS
Mb0863c|M.bovis_AF2122/97 DWDVVDRLADIAVPTLLVVGRFD-ECSPEHMREMQGRIAGSRLEFFESSS
MMAR_4789|M.marinum_M DWDVVDRLAEIALPTLLLAGKYD-ECSPEDMREMHQRIAGSRFEFFECSA
MUL_0358|M.ulcerans_Agy99 DWDVVDRLAEIALPTLLLAGKYD-ECSPEDMREMHQRIAGSRFEFFECSA
MAV_0868|M.avium_104 DWDVFDRLGEIAVPTLVLAGRYD-ECVPEHMWEMHRRIPGSRFELFESSA
MSMEG_2681|M.smegmatis_MC2_155 DWSVIDRLPDVTAPVLVIAGEHD-EATPKTWQPFVDHIPDVRSHVFPGTS
Mflv_2400|M.gilvum_PYR-GCK --------GAITAPTLVVWTSDDPTADVEEGRRIASMIPGARFEVMPGCG
TH_4528|M.thermoresistible__bu --------GAITAPTLVVWTSDDPTADVDEGKRIASMIPGARFEVMPGCG
Mvan_2912|M.vanbaalenii_PYR-1 ------LHFTLGLPTLLIWGDQDRLIPPAHGEAAHAALPGSRLVILPAVG
MAB_3934c|M.abscessus_ATCC_199 --------ADIKVPSLVIGFTDDMTLPPYLGKEVADAIPGGQYVEISDAG
MLBr_00862|M.leprae_Br4923 GRRRGKGVPVVNVPVQLIVNTMDRYVRPYGYDATARWVPRLWRRDIK-AG
: * :: * :. : .
Rv0840c|M.tuberculosis_H37Rv HMPFIEEPARFDRVMREFLRLHDI--------------------------
Mb0863c|M.bovis_AF2122/97 HMPFIEEPARFDRVMREFLRLHDI--------------------------
MMAR_4789|M.marinum_M HMPFIEEPDRFDRVMRDFLRSND---------------------------
MUL_0358|M.ulcerans_Agy99 HMPFIEEPDRFDRVMRDLLRSND---------------------------
MAV_0868|M.avium_104 HMPFIEEPEKFDAVMRDFLRLHD---------------------------
MSMEG_2681|M.smegmatis_MC2_155 HCTHLEKPEEFRAVVAQFLHQHDLAADARV--------------------
Mflv_2400|M.gilvum_PYR-GCK HWPQYEDPKTFDRLHLDFLLGRA---------------------------
TH_4528|M.thermoresistible__bu HWPQYEDPATFNRLHIDFLLGRS---------------------------
Mvan_2912|M.vanbaalenii_PYR-1 HFPQVEAPLAVADTLDDFIANSSPWHGPPRHADHAEAD------------
MAB_3934c|M.abscessus_ATCC_199 HLGFIERPDEVNRVILEFFASTAQTV------------------------
MLBr_00862|M.leprae_Br4923 HFSPMSHPQVMAAAVHDLADLVDGKQPSRALLRAQVGRSRDTFGDTLVSV
* . * . ::
Rv0840c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0863c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4789|M.marinum_M --------------------------------------------------
MUL_0358|M.ulcerans_Agy99 --------------------------------------------------
MAV_0868|M.avium_104 --------------------------------------------------
MSMEG_2681|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2400|M.gilvum_PYR-GCK --------------------------------------------------
TH_4528|M.thermoresistible__bu --------------------------------------------------
Mvan_2912|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3934c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 TGAGSGIGRETALALARRGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYA
Rv0840c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0863c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4789|M.marinum_M --------------------------------------------------
MUL_0358|M.ulcerans_Agy99 --------------------------------------------------
MAV_0868|M.avium_104 --------------------------------------------------
MSMEG_2681|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2400|M.gilvum_PYR-GCK --------------------------------------------------
TH_4528|M.thermoresistible__bu --------------------------------------------------
Mvan_2912|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3934c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LDVSDAAAVEAFAEQVSAKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVL
Rv0840c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0863c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4789|M.marinum_M --------------------------------------------------
MUL_0358|M.ulcerans_Agy99 --------------------------------------------------
MAV_0868|M.avium_104 --------------------------------------------------
MSMEG_2681|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2400|M.gilvum_PYR-GCK --------------------------------------------------
TH_4528|M.thermoresistible__bu --------------------------------------------------
Mvan_2912|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3934c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 AVNLGGVVNCCRAFGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKA
Rv0840c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0863c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4789|M.marinum_M --------------------------------------------------
MUL_0358|M.ulcerans_Agy99 --------------------------------------------------
MAV_0868|M.avium_104 --------------------------------------------------
MSMEG_2681|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2400|M.gilvum_PYR-GCK --------------------------------------------------
TH_4528|M.thermoresistible__bu --------------------------------------------------
Mvan_2912|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3934c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 ATYMFSDCLRAELDAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRD
Rv0840c|M.tuberculosis_H37Rv --------------------------------------------------
Mb0863c|M.bovis_AF2122/97 --------------------------------------------------
MMAR_4789|M.marinum_M --------------------------------------------------
MUL_0358|M.ulcerans_Agy99 --------------------------------------------------
MAV_0868|M.avium_104 --------------------------------------------------
MSMEG_2681|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_2400|M.gilvum_PYR-GCK --------------------------------------------------
TH_4528|M.thermoresistible__bu --------------------------------------------------
Mvan_2912|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3934c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 RRGQLDMMFRLRRYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVL
Rv0840c|M.tuberculosis_H37Rv -------------
Mb0863c|M.bovis_AF2122/97 -------------
MMAR_4789|M.marinum_M -------------
MUL_0358|M.ulcerans_Agy99 -------------
MAV_0868|M.avium_104 -------------
MSMEG_2681|M.smegmatis_MC2_155 -------------
Mflv_2400|M.gilvum_PYR-GCK -------------
TH_4528|M.thermoresistible__bu -------------
Mvan_2912|M.vanbaalenii_PYR-1 -------------
MAB_3934c|M.abscessus_ATCC_199 -------------
MLBr_00862|M.leprae_Br4923 PQVLRSTARIRVI