For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLTHPARRYLSSQAARPTGAFGRLLGRIWRAETADVNRIAVELLAPGPGERVCEIGFGPGRTLGLLAAAG AQVSGVEVSTTMIAIAAHHNAKAIAAGLISLYHGDGVTLPVADHSLDKVLGVHNFYFWPDPRASLCDIAR ALRPGGRLVLTSISDDQPLAARFDPAIYRVPPTLDTAAWLGAAGFIDVGIKRSADHPATVWFTATAT*
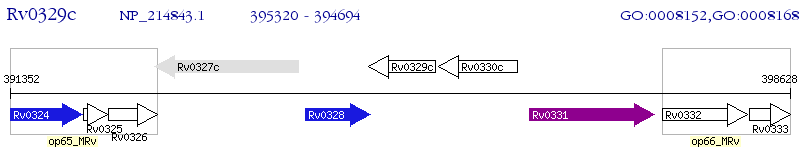
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0329c | - | - | 100% (208) | hypothetical protein Rv0329c |
| M. tuberculosis H37Rv | Rv3342 | - | 2e-09 | 31.67% (120) | methyltransferase (methylase) |
| M. tuberculosis H37Rv | Rv3729 | - | 4e-09 | 31.20% (125) | transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0336c | - | 1e-117 | 99.52% (208) | hypothetical protein Mb0336c |
| M. gilvum PYR-GCK | Mflv_4035 | - | 2e-12 | 31.34% (134) | methyltransferase type 11 |
| M. leprae Br4923 | MLBr_02273 | ubiE | 5e-08 | 29.77% (131) | ubiquinone/menaquinone biosynthesis methyltransferase |
| M. abscessus ATCC 19977 | MAB_3930c | - | 1e-08 | 30.26% (152) | ubiquinone/menaquinone biosynthesis methlytransferase UbiE |
| M. marinum M | MMAR_1183 | - | 1e-08 | 28.00% (150) | methyltransferase |
| M. avium 104 | MAV_2988 | - | 1e-09 | 27.01% (174) | ubiquinone/menaquinone biosynthesis methyltransferases |
| M. smegmatis MC2 155 | MSMEG_0098 | - | 8e-10 | 36.21% (116) | methyltransferase |
| M. thermoresistible (build 8) | TH_0087 | - | 1e-08 | 29.92% (127) | methyltransferase type 11 |
| M. ulcerans Agy99 | MUL_1436 | - | 8e-09 | 28.00% (150) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_2319 | - | 3e-10 | 34.51% (113) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02273|M.leprae_Br4923 MPPGSVDVVSRAALDKDPRDVVAMFDDVAHRYDLTNTV------------
MAB_3930c|M.abscessus_ATCC_199 --------MNRATLEKDPHEVASMFDAVARRYDITNTV------------
Rv0329c|M.tuberculosis_H37Rv --------MRLTHPARRYLSSQAARP--TGAFGRLLGR------------
Mb0336c|M.bovis_AF2122/97 --------MRLTHPARRYLSSQAARP--TGAFGRLLGR------------
Mflv_4035|M.gilvum_PYR-GCK -----------MDGSERPVNHHGDHPGFSGLTGQLCAV------------
Mvan_2319|M.vanbaalenii_PYR-1 -------------------------------------M------------
MMAR_1183|M.marinum_M --------------------MSGSAQDPSLSFGSAAAA------------
MUL_1436|M.ulcerans_Agy99 -------------------MVSGSAQDPSLSFGSAAAA------------
MAV_2988|M.avium_104 --------MGQVTDHWRAARLASALYDTAAMHDRVAAVGA----------
MSMEG_0098|M.smegmatis_MC2_155 MDTALRRAMDLLIDPPAEPDFSKGYLDLLGRGDSTAPKNTGLIQKAWASP
TH_0087|M.thermoresistible__bu --------------------------------------------------
MLBr_02273|M.leprae_Br4923 ----------LSLGQDRYWRRATRSALRIGPGQKVLDLAAGTAVSTAELS
MAB_3930c|M.abscessus_ATCC_199 ----------LSFGRDRFWRRATREALRLKPGDKVLDIAAGTAVSTAELT
Rv0329c|M.tuberculosis_H37Rv ----------IWRAETADVNRIAVELLAPGPGERVCEIGFGPGRTLGLLA
Mb0336c|M.bovis_AF2122/97 ----------IWRAETADVNRIAVELLAPGPGERVCEIGFGPGRTLGLLA
Mflv_4035|M.gilvum_PYR-GCK ----------AFLLTGRETGRLAADLTAVSATDHVLDIGCGPGNACRIAA
Mvan_2319|M.vanbaalenii_PYR-1 ----------LFLLAGRRTAGLAAALTEVTASDHVVDIGCGPGNGARIAA
MMAR_1183|M.marinum_M ----------YERGRPSYPPEAIDWLLPAGARD-VLDLGAGTGKLTTRLV
MUL_1436|M.ulcerans_Agy99 ----------YERGRPSYPPEAIDWLLPAGARD-VLDLGAGTGKLTTRLV
MAV_2988|M.avium_104 --------KALWGLALRDLLAEVRHVGAAPPGAYVLDIPCGGGFAFRGLR
MSMEG_0098|M.smegmatis_MC2_155 LGSKLYDRAQLLNRRLVASTRPPVEWLRIPPGGTVLDIGCGPGNITAQLA
TH_0087|M.thermoresistible__bu ------------------MLTVDYDRLGIGPGVSVIDVGCGLGRHSFEAY
. * :: * .
MLBr_02273|M.leprae_Br4923 K-SGAWCVAADF------SVRMLATGG--------ARKVPKVAADATQLP
MAB_3930c|M.abscessus_ATCC_199 Q-SGAWCVAADF------SVGMLKAGA--------RRDVPKVAGDATRLP
Rv0329c|M.tuberculosis_H37Rv A-AGAQVSGVEV------STTMIAIAAHHNAKAIAAGLISLYHGDGVTLP
Mb0336c|M.bovis_AF2122/97 A-AGAQVSGVEV------STTMIAIAAHHNAKAIAAGLISLYHGDGVTLP
Mflv_4035|M.gilvum_PYR-GCK A-RGARVTGVDP------SAAMLRVAR---IVTHG-ASVTWVRGGAEALP
Mvan_2319|M.vanbaalenii_PYR-1 Q-RGARVTGVDP------SRSMLRVAR---AVTRGRPAITWAEGTAEALP
MMAR_1183|M.marinum_M E-RGLDVVAVDP------IPEMLEVLR------ASLPRTLALLGTAEEIP
MUL_1436|M.ulcerans_Agy99 E-RGLDVVAVDP------IPEMLEVLR------ASLPRTLALLGTAEEIP
MAV_2988|M.avium_104 AGQDCRYVAADI------SSDMLRRARSRATRFGVANLMEFTEADITCLP
MSMEG_0098|M.smegmatis_MC2_155 RAAGLDGLALGV------DISEPMLAR--AVAAEAGRQVGFVRADAQQLP
TH_0087|M.thermoresistible__bu R-RGAHIVAFDQNAEELNEVDTILQAMAEQREAPAGARAEVVKGDALDLP
. . . :*
MLBr_02273|M.leprae_Br4923 FSDGVFDAVTISFGLRNIADYQAALREMARVTRPGGQLVVCEFSTPTNAL
MAB_3930c|M.abscessus_ATCC_199 FADHSFDAVTISFGLRNVVDHVEGLREMARVTKPGGRLAVCEFSTPVNRP
Rv0329c|M.tuberculosis_H37Rv VADHSLDKVLGVHNFYFWPDPRASLCDIARALRPGGRLVLTSIS------
Mb0336c|M.bovis_AF2122/97 VADHSLDKVLGVHNFYFSPDPRASLCDIARALRPGGRLVLTSIS------
Mflv_4035|M.gilvum_PYR-GCK VPDAGATVAWALATVHHWPDVDAGLAEIHRALGPGARFLAVER-------
Mvan_2319|M.vanbaalenii_PYR-1 VPDASATVVWALATVHHWRDVGAGLSEIHRVLVPGGRLLAVER-------
MMAR_1183|M.marinum_M LEDNSVDAVLVAQ-AWHWVDPARAIPEVARVLRPGGRLGLVWNTRDERLG
MUL_1436|M.ulcerans_Agy99 LEDNSVDAVLVAQ-AWHWVDPARAIPEVARVLRPGGRLGLVWNTRDERLG
MAV_2988|M.avium_104 FPDNTFDLALTFNGLHCLPDPRAAVFELARVLKPGGILRGTTCVRGRG--
MSMEG_0098|M.smegmatis_MC2_155 FRDEVFDAATSLAVFQLIPDPVAAVSEIVRVLKPGGRVAIMVPTAG----
TH_0087|M.thermoresistible__bu YADGTFDRVIASEILEHVPEDDRAIAELVRVLKPGGKLVVTVPRWLPERI
* . : .: :: *. **. .
MLBr_02273|M.leprae_Br4923 VANVYTEYLMRALPQVARLVSSNP---DAYIYLAESIRAWPDQVTLACQL
MAB_3930c|M.abscessus_ATCC_199 FRTLYMEYLMKTLPEVARAVSSNP---DAYVYLAESIRAWPDQEALAHQI
Rv0329c|M.tuberculosis_H37Rv ----------DDQPLAARFDP--------------AIYRVPPTLDTAAWL
Mb0336c|M.bovis_AF2122/97 ----------DDQPLAARFDP--------------AIYRVPPTLDTAAWL
Mflv_4035|M.gilvum_PYR-GCK ----------QTEPDATGISS--------------HGWTGAQAETFAAMC
Mvan_2319|M.vanbaalenii_PYR-1 ----------QVAPDATGFAS--------------HGWTGQQAETFAALC
MMAR_1183|M.marinum_M WVRELGRIIGRDSDPVRDKVTLP----EPFTGVARHHVEWTNYLTPQALI
MUL_1436|M.ulcerans_Agy99 WVRELGRIIGRDSDPVRDKVTLP----EPFTGVARHHVEWTNYLTPQALI
MAV_2988|M.avium_104 ---------WRQDRFVTALQAAG------------FFGNTPGAGDVSRWL
MSMEG_0098|M.smegmatis_MC2_155 -----------AVKPVTFLARGG--------------ARIFGDDELGDIF
TH_0087|M.thermoresistible__bu CWLLSDEYHANEGGHVRIYRADELRDKITRHGLRLTHRHHAHALHSPFWW
.
MLBr_02273|M.leprae_Br4923 SRTG------------------------------WASPRWRNLTGGIVAL
MAB_3930c|M.abscessus_ATCC_199 ASAG------------------------------WSNVRWRNLTGGIVAL
Rv0329c|M.tuberculosis_H37Rv GAAG------------------------------FIDVGIKRSADHPATV
Mb0336c|M.bovis_AF2122/97 GAAG------------------------------FIDVGIKRSADHPATV
Mflv_4035|M.gilvum_PYR-GCK ESAG------------------------------LTTVRVGSGRAGKRRV
Mvan_2319|M.vanbaalenii_PYR-1 VTAG------------------------------LADAKVNADRAGRREV
MMAR_1183|M.marinum_M DLVAS-------RSYCITSPAEVRTKTLDQVRELLATHPALANSAGLALP
MUL_1436|M.ulcerans_Agy99 DLVAS-------RSYCITSPAEVRTKTLDQVRELLATHPALANSAGLALP
MAV_2988|M.avium_104 GEAG--------------------------------LEIVAARHSGSVEL
MSMEG_0098|M.smegmatis_MC2_155 ENVG--------------------------------LVGVRVKTHGFIQW
TH_0087|M.thermoresistible__bu LKCAVGTNNDRHFAVRAYHQLLVWDMVRRPWVTRWAEEMLNPLIGKSVAL
.
MLBr_02273|M.leprae_Br4923 HAADKPVR----
MAB_3930c|M.abscessus_ATCC_199 HIATL-------
Rv0329c|M.tuberculosis_H37Rv WFTATAT-----
Mb0336c|M.bovis_AF2122/97 WFTATAT-----
Mflv_4035|M.gilvum_PYR-GCK WTVHATRP----
Mvan_2319|M.vanbaalenii_PYR-1 WTVHAVRP----
MMAR_1183|M.marinum_M YVTVCVSATLAA
MUL_1436|M.ulcerans_Agy99 YVTVCVSATLAA
MAV_2988|M.avium_104 FEAVRPSA----
MSMEG_0098|M.smegmatis_MC2_155 VRGTKL------
TH_0087|M.thermoresistible__bu YFDKPEVAGADS