For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVPDQVAGPTPARTSRATAIEVDDLRLRYRGASTNVLAGVNLRVEHGETVALIGTNGAGKSTLLRCLVRL AEPTSGTVTLAGTDVTAAPRRALRGIRRDIGFVFQRFHLIPRLTVVHNVVHGAMGRHGTRCAWPLTSPAE VRREAMASLDRVGLASLAERRVDTLSGGQQQRVAIARMLMQRPTIILADEPVASLDPASATAVMELLSGI AAERHVTVVMALHQMDLAMSYADRVVGLRGGTVDFDKPAAECDAAQLEPIFADGVS*
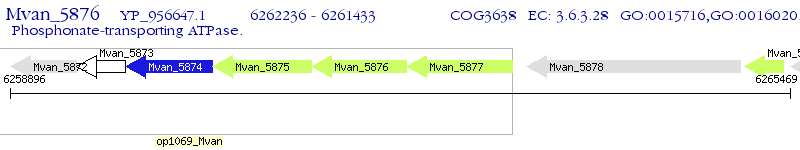
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5876 | - | - | 100% (267) | phosphonate ABC transporter, ATPase subunit |
| M. vanbaalenii PYR-1 | Mvan_2905 | - | 1e-57 | 54.09% (220) | phosphonate ABC transporter, ATPase subunit |
| M. vanbaalenii PYR-1 | Mvan_4704 | - | 1e-34 | 37.45% (243) | ABC transporter-related protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1012 | - | 2e-32 | 36.97% (238) | adhesion component transport ATP-binding protein ABC |
| M. gilvum PYR-GCK | Mflv_1083 | - | 3e-36 | 38.10% (252) | ABC transporter related |
| M. tuberculosis H37Rv | Rv0986 | - | 2e-32 | 36.97% (238) | adhesion component transport ATP-binding protein ABC |
| M. leprae Br4923 | MLBr_00669 | ftsE | 8e-33 | 37.39% (230) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_4237c | - | 5e-36 | 39.11% (248) | amino acid ABC transporter ATP-binding protein |
| M. marinum M | MMAR_0060 | - | 1e-34 | 42.41% (224) | glutamine ABC transporter, ATP-binding protein |
| M. avium 104 | MAV_0058 | - | 5e-34 | 42.67% (225) | ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_0647 | phnC | 3e-57 | 54.09% (220) | phosphonate ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4568 | - | 2e-60 | 54.74% (232) | PUTATIVE phosphonate ABC transporter, ATP-binding protein |
| M. ulcerans Agy99 | MUL_0674 | - | 5e-32 | 40.38% (213) | ABC transporter, ATP-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0647|M.smegmatis_MC2_155 ----------MNPVAGDDVVVIARDVTKRFG--DTLALDHVSLDVHRSEL
TH_4568|M.thermoresistible__bu -----LTAPPVEPVAGDDLVIAVRDATKRFG--GTVALDGVSLDVHRSEF
Mvan_5876|M.vanbaalenii_PYR-1 MVVPDQVAGPTPARTSRATAIEVDDLRLRYRGASTNVLAGVNLRVEHGET
MMAR_0060|M.marinum_M --------MTRQDAGRQPVCLAARSIHQTLG--GSVVLRGVDLEVPAGTT
MAV_0058|M.avium_104 --------MS-NPASRASVSLACKDIHQTLN--GNAVLRGVDLDTPAGST
MAB_4237c|M.abscessus_ATCC_199 --------MS---------TIEVRGVRKSYG--QVTALRDVNLTVHQGEV
Mflv_1083|M.gilvum_PYR-GCK -------------MTNRGVGVQFDELTRVYG--ATRALDGLTLHIEPGEL
MLBr_00669|M.leprae_Br4923 ------------MITLDHVSKKYKSLARP-------ALDNVNVKIDKGEF
Mb1012|M.bovis_AF2122/97 -----MNRQPI-------VQLSNLSWTFREGETRRQVLDHITFDFEPGEF
Rv0986|M.tuberculosis_H37Rv -----MNRQPI-------VQLSNLSWTFREGETRRQVLDHITFDFEPGEF
MUL_0674|M.ulcerans_Agy99 -----MTRAQAPTSPRGGVSLHGVDKTFGTAAASVHALRRIDLDIEPGEL
.* : . .
MSMEG_0647|M.smegmatis_MC2_155 LVLLGLSGSGKSTLLRCLNGLHPVTSGTVDVGGT---------RVDQASG
TH_4568|M.thermoresistible__bu VVLLGLSGSGKSTLLRCLNGLHRVTSGTVEVGGV---------RVNTASR
Mvan_5876|M.vanbaalenii_PYR-1 VALIGTNGAGKSTLLRCLVRLAEPTSGTVTLAGT---------DVTAAPR
MMAR_0060|M.marinum_M AAVIGPSGSGKSTLLRTLNRLHEPDSGDILLDGR--------SVLRDNPN
MAV_0058|M.avium_104 VAVIGPSGSGKSTLLRTLNRLYEPDRGDILLDGR--------SVLHDDPD
MAB_4237c|M.abscessus_ATCC_199 TVIIGPSGSGKSTLLRALNHLEKVDAGTIRIDGELIGYRQRGSVLYELPE
Mflv_1083|M.gilvum_PYR-GCK VALLGPSGGGKTTALRILAGLDEATSGTVSVGGV---------DVGRVPA
MLBr_00669|M.leprae_Br4923 VFLIGPSGSGKSTFMRLLLGAETPTSGDVRVSKF---------HVNKLPG
Mb1012|M.bovis_AF2122/97 VALLGQSGSGKSTLLNLISGIEKPTTGDVTINGFA--------ITQKTER
Rv0986|M.tuberculosis_H37Rv VALLGQSGSGKSTLLNLISGIEKPTTGDVTINGFA--------ITQKTER
MUL_0674|M.ulcerans_Agy99 VAIVGESGSGKTTLLNVIGGIEEANSGSINVAGQD--------ISGCRPR
::* .*.**:* :. : * : :
MSMEG_0647|M.smegmatis_MC2_155 AQLRALRRRVGFVFQHFNLVGRLSCLENVLIGGLGRLRLPRYGALTYPRH
TH_4568|M.thermoresistible__bu RELRRLRRRVGFVFQQFNLVGRLSCLENVLIGGLGRLTLPRYGALTYPRR
Mvan_5876|M.vanbaalenii_PYR-1 RALRGIRRDIGFVFQRFHLIPRLTVVHNVVHGAMGRHGTRCAWPLTSPAE
MMAR_0060|M.marinum_M D--LRQR--IGMVFQHFNLFPHRNVVDNVTLAPRKLRGLSNE-------E
MAV_0058|M.avium_104 Q--LRQR--IGMVFQHFNLFPHRTVLDNVALAPRKLRRLPAE-------A
MAB_4237c|M.abscessus_ATCC_199 RAILRQRSQVGFVFQNFNLFPHLTVLENVIEAPLAARRAPRA-------E
Mflv_1083|M.gilvum_PYR-GCK N-----KRDMGMVFQAYSLFPHLTVLDNVAFGLKMRGKGKSD-------R
MLBr_00669|M.leprae_Br4923 RHIPRLRQVIGCVFQDFRLLQQKTVYENVAFALEVIGR-RSD-------A
Mb1012|M.bovis_AF2122/97 DRTLFRRDQIGIVFQFFNLIPTLTVLENITLPQELAGVSQRKAA------
Rv0986|M.tuberculosis_H37Rv DRTLFRRDQIGIVFQFFNLIPTLTVLENITLPQELAGVSQRKAA------
MUL_0674|M.ulcerans_Agy99 ALSKFRRDHVGFIFQFFNLIPTLTAKENVEVIVELTGRGDRRR-------
: :* :** : *. . .*:
MSMEG_0647|M.smegmatis_MC2_155 MRAEALAHLDRVGLADYADRRADTLSGGQQQRVAIARTLMQKPALLLADE
TH_4568|M.thermoresistible__bu MRTEALEHLDRVGLAGLAEQRADTLSGGQQQRVAIARTLMQRPSLLLADE
Mvan_5876|M.vanbaalenii_PYR-1 VRREAMASLDRVGLASLAERRVDTLSGGQQQRVAIARMLMQRPTIILADE
MMAR_0060|M.marinum_M ARMLALDQLDRVGLKHKANARPDMLSGGQQQRVAIARALAMAPQVMFFDE
MAV_0058|M.avium_104 ARELALTQLDRVGLKHKAGARPATLSGGQQQRVAIARALAMSPQVMFFDE
MAB_4237c|M.abscessus_ATCC_199 LEAEATRLLERVGVADKAAQYPRRLSGGQQQRVAIARALALRPKVILFDE
Mflv_1083|M.gilvum_PYR-GCK LSRAA-DMLDLVGLGELGGRYANELSGGQQQRVALARALAIRPRVLLLDE
MLBr_00669|M.leprae_Br4923 INQVVPDVLETVGLSGKANRLPDELSGGEQQRVAIARAFVNRPLVLLADE
Mb1012|M.bovis_AF2122/97 --VVARDLLEKVGMADRERTFPDKLSGGEQQRVAISRALAHNPMLVLADE
Rv0986|M.tuberculosis_H37Rv --VVARDLLEKVGMADRERTFPDKLSGGEQQRVAISRALAHNPMLVLADE
MUL_0674|M.ulcerans_Agy99 ----ATELLDAVELADRAGSFPGQLSGGQQQRVAIARALITNPDLLLADE
. *: * : ****:*****::* : * ::: **
MSMEG_0647|M.smegmatis_MC2_155 PVASLDPENAGVVMDLLFRVCIEEKLTVVCTLHQVDLALGWAHRLVGLQG
TH_4568|M.thermoresistible__bu PVASLDPENAGVVMDLLFQVCMEQKLTVVCTLHQIDLALGWAHRLVGLRA
Mvan_5876|M.vanbaalenii_PYR-1 PVASLDPASATAVMELLSGIAAERHVTVVMALHQMDLAMSYADRVVGLRG
MMAR_0060|M.marinum_M ATSALDPEMVKGILALIADLGADG-MTMVVVTHEMGFARSTSDAVVFMDH
MAV_0058|M.avium_104 ATSALDPEMVKGILQLIADLGAEG-MTMVVVTHEMGFARSASDAVIFMDH
MAB_4237c|M.abscessus_ATCC_199 PTSALDPELVTEVLDVIAQLARDG-ATLVIVTHEIGLAREIADTVVFMDR
Mflv_1083|M.gilvum_PYR-GCK PLSALDAKVRTQLRDEIRRVQLEVGTTTLFVTHDQEEALAVADRVGVMNQ
MLBr_00669|M.leprae_Br4923 PTGNLDPDTSKDIMDLLERIN-RTGTTVLMATHDHHIVDSMRQRVVELSL
Mb1012|M.bovis_AF2122/97 PTGNLDSDTGDKVLDVLLDLTRQAGKTLIMATHS-PSMTQHADRVVNLQG
Rv0986|M.tuberculosis_H37Rv PTGNLDSDTGDKVLDVLLDLTRQAGKTLIMATHS-PSMTQHADRVVNLQG
MUL_0674|M.ulcerans_Agy99 PMGALDVATGRRVLELLQQTNR-SGLGVIMVTHN-RAAADVADPIIQMRD
. . ** : : : . *. . : :
MSMEG_0647|M.smegmatis_MC2_155 GRKVLDRPAVGMTRDDVMAVYQRVEPAVT--PARRV--------------
TH_4568|M.thermoresistible__bu GRKVLDRPAVGASHDDVLDIYRRVDPAVIGSPAGR---------------
Mvan_5876|M.vanbaalenii_PYR-1 GTVDFDKPAAECDAAQLEPIFADGVS------------------------
MMAR_0060|M.marinum_M GTVVEAGPPEQLFEAARTERLQRFLAQVL---------------------
MAV_0058|M.avium_104 GKVVESGPPEQIFEAAETERLQRFLSQVL---------------------
MAB_4237c|M.abscessus_ATCC_199 GAIVEQGPPKEVLDSPSHPRTASFLSKVLS--------------------
Mflv_1083|M.gilvum_PYR-GCK GRLEQLAAPADLYAHPATPFVADFVGLNNKVPASVSGGTATLLGVSVPAL
MLBr_00669|M.leprae_Br4923 GRLVR----------------DEWCGIYG---------------------
Mb1012|M.bovis_AF2122/97 GRLIPALNRENQTDQPASTILLPTSYE-----------------------
Rv0986|M.tuberculosis_H37Rv GRLIPAVNRENQTDQPASTILLPTSYE-----------------------
MUL_0674|M.ulcerans_Agy99 GAIVE--QRRNSAPAEASSVRW----------------------------
*
MSMEG_0647|M.smegmatis_MC2_155 --------------------------------------------------
TH_4568|M.thermoresistible__bu --------------------------------------------------
Mvan_5876|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0060|M.marinum_M --------------------------------------------------
MAV_0058|M.avium_104 --------------------------------------------------
MAB_4237c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_1083|M.gilvum_PYR-GCK PGSVDGDGVAMVRPEAVTVAADPAGTATVATVSFLGAITRVGITMPDGSL
MLBr_00669|M.leprae_Br4923 ----------MDR-------------------------------------
Mb1012|M.bovis_AF2122/97 --------------------------------------------------
Rv0986|M.tuberculosis_H37Rv --------------------------------------------------
MUL_0674|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_0647|M.smegmatis_MC2_155 ----------------------------------
TH_4568|M.thermoresistible__bu ----------------------------------
Mvan_5876|M.vanbaalenii_PYR-1 ----------------------------------
MMAR_0060|M.marinum_M ----------------------------------
MAV_0058|M.avium_104 ----------------------------------
MAB_4237c|M.abscessus_ATCC_199 ----------------------------------
Mflv_1083|M.gilvum_PYR-GCK LSAQMSSTAARALTPGDPVTVGVEPGGVLVTRPL
MLBr_00669|M.leprae_Br4923 ----------------------------------
Mb1012|M.bovis_AF2122/97 ----------------------------------
Rv0986|M.tuberculosis_H37Rv ----------------------------------
MUL_0674|M.ulcerans_Agy99 ----------------------------------