For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STIEVRGVRKSYGQVTALRDVNLTVHQGEVTVIIGPSGSGKSTLLRALNHLEKVDAGTIRIDGELIGYRQ RGSVLYELPERAILRQRSQVGFVFQNFNLFPHLTVLENVIEAPLAARRAPRAELEAEATRLLERVGVADK AAQYPRRLSGGQQQRVAIARALALRPKVILFDEPTSALDPELVTEVLDVIAQLARDGATLVIVTHEIGLA REIADTVVFMDRGAIVEQGPPKEVLDSPSHPRTASFLSKVLS*
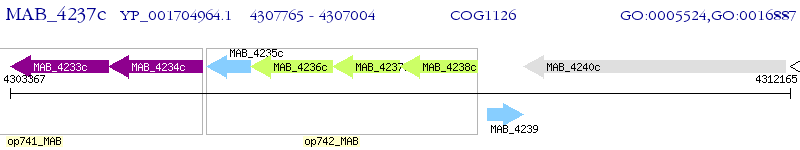
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4237c | - | - | 100% (253) | amino acid ABC transporter ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_3050 | - | 1e-67 | 51.39% (251) | putative glutamate ABC transporter, ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_1655 | - | 4e-45 | 39.84% (251) | sulfate ABC transporter, ATP-binding protein SysA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2419c | cysA1 | 5e-43 | 39.76% (249) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. gilvum PYR-GCK | Mflv_3581 | - | 8e-80 | 58.20% (244) | ABC transporter related |
| M. tuberculosis H37Rv | Rv2397c | cysA1 | 5e-43 | 39.76% (249) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. leprae Br4923 | MLBr_01089 | - | 4e-36 | 36.00% (250) | putative ABC-transport protein, ATP-binding component |
| M. marinum M | MMAR_0060 | - | 2e-57 | 49.40% (249) | glutamine ABC transporter, ATP-binding protein |
| M. avium 104 | MAV_0058 | - | 4e-59 | 48.80% (250) | ABC transporter, ATP-binding protein |
| M. smegmatis MC2 155 | MSMEG_0178 | - | 2e-80 | 57.26% (248) | ATP-binding protein |
| M. thermoresistible (build 8) | TH_1784 | - | 2e-71 | 54.22% (249) | ABC transporter, ATP-binding protein GluA |
| M. ulcerans Agy99 | MUL_3658 | cysA1 | 3e-42 | 39.76% (249) | sulfate-transport ATP-binding protein ABC transporter CysA1 |
| M. vanbaalenii PYR-1 | Mvan_2834 | - | 1e-78 | 56.97% (244) | ABC transporter-related protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2419c|M.bovis_AF2122/97 ----------MT------YAIVVADATKRYG-DFVALDHVDFVVPTGSLT
Rv2397c|M.tuberculosis_H37Rv ----------MT------YAIVVADATKRYG-DFVALDHVDFVVPTGSLT
MUL_3658|M.ulcerans_Agy99 ----------MTGPGTGGHAIVVRDAFKQYG-DFVALDHVDFVVPAGSLT
Mflv_3581|M.gilvum_PYR-GCK ---------MTTSENSAHPMVKAELVCKDFG-ALKVLKGITLEVQKGQVL
Mvan_2834|M.vanbaalenii_PYR-1 ---------MTADAVSAKPMVKAELVCKDFG-ALKVLKGITLEVQKGQVL
MSMEG_0178|M.smegmatis_MC2_155 ---------MTVPSETARPLVRAQSVTKSFG-TVRVLDNVDVEIARGEVV
MAB_4237c|M.abscessus_ATCC_199 ---------MST--------IEVRGVRKSYG-QVTALRDVNLTVHQGEVT
MMAR_0060|M.marinum_M --------MTRQDAGRQPVCLAARSIHQTLG-GSVVLRGVDLEVPAGTTA
MAV_0058|M.avium_104 --------MS-NPASRASVSLACKDIHQTLN-GNAVLRGVDLDTPAGSTV
TH_1784|M.thermoresistible__bu MDSAGADVAVADPAAEAVPMISMRGVNKYFG-SLHVLRDINLDVARGQVV
MLBr_01089|M.leprae_Br4923 -----------------MAEIVLEHVNKNYPNGATAVHDLSITVADGEFL
: : .: : . *
Mb2419c|M.bovis_AF2122/97 ALLGPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVP-----------
Rv2397c|M.tuberculosis_H37Rv ALLGPSGSGKSTLLRTIAGLDQPDTGTITINGRDVTRVP-----------
MUL_3658|M.ulcerans_Agy99 ALLGPSGSGKSTLLRTIAGLDQPDSGNITINGRDVTRVP-----------
Mflv_3581|M.gilvum_PYR-GCK VLVGPSGSGKSTLLRCINHLETVSAGRLYVDGTLVGYHERGGKLYEMKPR
Mvan_2834|M.vanbaalenii_PYR-1 VLVGPSGSGKSTFLRCINHLETVSAGRLYVDGALVGYNERGGKLHEMKPR
MSMEG_0178|M.smegmatis_MC2_155 CLIGPSGSGKSTFLRCINHLETLDGGQLWVDGELVGYRRKGDKLIELSDR
MAB_4237c|M.abscessus_ATCC_199 VIIGPSGSGKSTLLRALNHLEKVDAGTIRIDGELIGYRQRGSVLYELPER
MMAR_0060|M.marinum_M AVIGPSGSGKSTLLRTLNRLHEPDSGDILLDGRSVLRD------------
MAV_0058|M.avium_104 AVIGPSGSGKSTLLRTLNRLYEPDRGDILLDGRSVLHD------------
TH_1784|M.thermoresistible__bu VVLGPSGSGKSTLCRTLNRLEPIDSGAISIDGVALPAEG----------R
MLBr_01089|M.leprae_Br4923 ILIGPSGCGKTTTLNMIAGLEDISSGELRIDGDRVNEKAP----------
::****.**:* . : * . * : ::* :
Mb2419c|M.bovis_AF2122/97 ---PQRRGIGFVFQHYAAFKHLTVRDNVAFG-LKIRKRPKAEIKAKVDNL
Rv2397c|M.tuberculosis_H37Rv ---PQRRGIGFVFQHYAAFKHLTVRDNVAFG-LKIRKRPKAEIKAKVDNL
MUL_3658|M.ulcerans_Agy99 ---PQRRGIGFVFQHYAAFKHLTVRDNVAYG-LKIRKRPKAEIRDKVDNL
Mflv_3581|M.gilvum_PYR-GCK DVARQRRDVGMVFQHFNLFPHRTALGNVVEAPIQVKGVKKDEAVTRGREL
Mvan_2834|M.vanbaalenii_PYR-1 DVAKQRRDVGMVFQHFNLFPHRTALGNVVEAPIQVKGVRKAQAIEKGKEL
MSMEG_0178|M.smegmatis_MC2_155 QASVRRAEIGMVFQHFNLFPHMTVIQNIVEAPVRTRRATLEEATARGEDL
MAB_4237c|M.abscessus_ATCC_199 AILRQRSQVGFVFQNFNLFPHLTVLENVIEAPLAARRAPRAELEAEATRL
MMAR_0060|M.marinum_M NPNDLRQRIGMVFQHFNLFPHRNVVDNVTLAPRKLRGLSNEEARMLALDQ
MAV_0058|M.avium_104 DPDQLRQRIGMVFQHFNLFPHRTVLDNVALAPRKLRRLPAEAARELALTQ
TH_1784|M.thermoresistible__bu ELARLRADVGMVFQQFNLFAHKTIVDNVALGPMKVRKTPREQARAEAMAL
MLBr_01089|M.leprae_Br4923 ----KDRDIAMVFQSYALYPHMTVRQNIAFP-LMLAKVKKAEIAQKVSET
:.:*** : : * . *:
Mb2419c|M.bovis_AF2122/97 LQVVGLSGFQSRYPNQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAK
Rv2397c|M.tuberculosis_H37Rv LQVVGLSGFQSRYPNQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAK
MUL_3658|M.ulcerans_Agy99 LEVVGLSGFHGRYPDQLSGGQRQRMALARALAVDPEVLLLDEPFGALDAK
Mflv_3581|M.gilvum_PYR-GCK LAQVGLADKADAYPAQLSGGQQQRVAIARALAMDPKLMLFDEPTSALDPE
Mvan_2834|M.vanbaalenii_PYR-1 LAQVGLADKAGAYPAQLSGGQQQRVAIARALAMDPKLMLFDEPTSALDPE
MSMEG_0178|M.smegmatis_MC2_155 LTRMGLADKRTVYPAELSGGQKQRVAIARALAMQPKLMLFDEPTSALDPE
MAB_4237c|M.abscessus_ATCC_199 LERVGVADKAAQYPRRLSGGQQQRVAIARALALRPKVILFDEPTSALDPE
MMAR_0060|M.marinum_M LDRVGLKHKANARPDMLSGGQQQRVAIARALAMAPQVMFFDEATSALDPE
MAV_0058|M.avium_104 LDRVGLKHKAGARPATLSGGQQQRVAIARALAMSPQVMFFDEATSALDPE
TH_1784|M.thermoresistible__bu LERVGVADQADKYPAQLSGGQQQRVAIARSLAMNPKVMLFDEPTSALDPE
MLBr_01089|M.leprae_Br4923 AQILDLTDLLDRKPSQLSGGQRQRVAMGRAIVRHPKAFLMDEPLSNLDAK
:.: * *****:**:*:.*::. *: :::**. . **.:
Mb2419c|M.bovis_AF2122/97 VREELRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGS
Rv2397c|M.tuberculosis_H37Rv VREELRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLHKGRIEQVGS
MUL_3658|M.ulcerans_Agy99 VREDLRAWLRRLHDEVHVTTVLVTHDQAEALDVADRIAVLNSGRIEQVGS
Mflv_3581|M.gilvum_PYR-GCK LVGEVLAVMKKLAAQG-MTMVVVTHEMGFAREVADELVFMDEGVVVERGN
Mvan_2834|M.vanbaalenii_PYR-1 LVGEVLGVMKKLAAQG-MTMVVVTHEMGFAREVADELVFMDAGVVVERGA
MSMEG_0178|M.smegmatis_MC2_155 LVGDVLRVMKGLAESG-MTMIIVTHEIGFAREVADRVIFFENGRVVEQGS
MAB_4237c|M.abscessus_ATCC_199 LVTEVLDVIAQLARDG-ATLVIVTHEIGLAREIADTVVFMDRGAIVEQGP
MMAR_0060|M.marinum_M MVKGILALIADLGADG-MTMVVVTHEMGFARSTSDAVVFMDHGTVVEAGP
MAV_0058|M.avium_104 MVKGILQLIADLGAEG-MTMVVVTHEMGFARSASDAVIFMDHGKVVESGP
TH_1784|M.thermoresistible__bu MINEVLAVMSALAADG-MTMLVVTHEMGFARRASDRVVFMSDGAIVEDAE
MLBr_01089|M.leprae_Br4923 LRVRTRGEIARLQRRLGATTVYVTHDQTEAMTLGDRVVVMRSGVVQQIGT
: : * * : ***: * .* : .: * : : .
Mb2419c|M.bovis_AF2122/97 PTDVYDAPANAFVMSFLGAVSTLNGSLVRPHD------------------
Rv2397c|M.tuberculosis_H37Rv PTDVYDAPANAFVMSFLGAVSTLNGSLVRPHD------------------
MUL_3658|M.ulcerans_Agy99 PTEVYDAPTNAFVMSFLGAVSALNGALVRPHD------------------
Mflv_3581|M.gilvum_PYR-GCK PREVMSNPQHERTKAFLSKVM-----------------------------
Mvan_2834|M.vanbaalenii_PYR-1 PREVMANPQHERTKAFLSKVM-----------------------------
MSMEG_0178|M.smegmatis_MC2_155 ARDVLDSPSNPRTAAFLSAVR-----------------------------
MAB_4237c|M.abscessus_ATCC_199 PKEVLDSPSHPRTASFLSKVLS----------------------------
MMAR_0060|M.marinum_M PEQLFEAARTERLQRFLAQVL-----------------------------
MAV_0058|M.avium_104 PEQIFEAAETERLQRFLSQVL-----------------------------
TH_1784|M.thermoresistible__bu PARFFEDPQTDRARDFLGKILHH---------------------------
MLBr_01089|M.leprae_Br4923 PDELYERPVNLFVAGFIGSPVMNFFPAKLTPNGLTLPLGELNLSRDVQAM
. . . *:.
Mb2419c|M.bovis_AF2122/97 IRVGRTPNMAVAAADGTAGSTGVLRAVVDRVVVLGFEVRVELTSAATGGA
Rv2397c|M.tuberculosis_H37Rv IRVGRTPNMAVAAADGTAGSTGVLRAVVDRVVVLGFEVRVELTSAATGGA
MUL_3658|M.ulcerans_Agy99 IRVGRTPEMAVAAADGTGESTGVTRAVVDRIVVLGFEVRVELTSAATGGA
Mflv_3581|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2834|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0178|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4237c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0060|M.marinum_M --------------------------------------------------
MAV_0058|M.avium_104 --------------------------------------------------
TH_1784|M.thermoresistible__bu --------------------------------------------------
MLBr_01089|M.leprae_Br4923 IGQHPVPDRVIVGVRPEHLTDAKLIDAHQHGTVLTFKVKVDLVESLGADK
Mb2419c|M.bovis_AF2122/97 FTAQITRGDAEALALREGDTVYVRATRVPPIAGGVSGVDDAGVERVKVTS
Rv2397c|M.tuberculosis_H37Rv FTAQITRGDAEALALREGDTVYVRATRVPPIAGGVSGVDDAGVERVKVTS
MUL_3658|M.ulcerans_Agy99 FTAQITRGDAEALALQEGDTVYVRATRVPPIAGAAPAVPDDGAGSTPRAA
Mflv_3581|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2834|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0178|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4237c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_0060|M.marinum_M --------------------------------------------------
MAV_0058|M.avium_104 --------------------------------------------------
TH_1784|M.thermoresistible__bu --------------------------------------------------
MLBr_01089|M.leprae_Br4923 YIYFTTTGCDVYSAQLDELASELEVRENQFVARVSAESKMAIGESIELAF
Mb2419c|M.bovis_AF2122/97 T-----------------------
Rv2397c|M.tuberculosis_H37Rv T-----------------------
MUL_3658|M.ulcerans_Agy99 QIGVGGQ-----------------
Mflv_3581|M.gilvum_PYR-GCK ------------------------
Mvan_2834|M.vanbaalenii_PYR-1 ------------------------
MSMEG_0178|M.smegmatis_MC2_155 ------------------------
MAB_4237c|M.abscessus_ATCC_199 ------------------------
MMAR_0060|M.marinum_M ------------------------
MAV_0058|M.avium_104 ------------------------
TH_1784|M.thermoresistible__bu ------------------------
MLBr_01089|M.leprae_Br4923 GTAKIAVFDADSGVNLTVSAPTDA