For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPASRPADHDVAGDDVVIAAHNVTKRFGNTLALDDVSLEVRRSELLVLLGLSGSGKSTLLRCLNGLHPVT SGTVEVDGTRVDGASKAQLRALRRNVGFVFQHFNLVGRLSCLENVLIGGLGQLTFPRYGALTYPKRMRTE ALAHLDRVGLADFAERRADTLSGGQQQRVAIARTLMQAPALLLADEPVASLDPENAGLVMDLLFRVCIEE KLTVVCTLHQVDLALGWAHRLVGLRNGRKVLDRPAVGLTRDEVMGIYQRVDPAVDSAPRSAVT
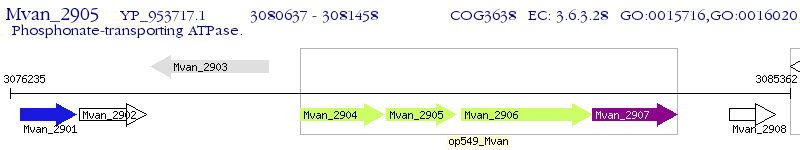
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2905 | - | - | 100% (273) | phosphonate ABC transporter, ATPase subunit |
| M. vanbaalenii PYR-1 | Mvan_5876 | - | 1e-57 | 54.09% (220) | phosphonate ABC transporter, ATPase subunit |
| M. vanbaalenii PYR-1 | Mvan_2428 | - | 2e-34 | 38.86% (229) | ABC transporter-related protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3129c | ftsE | 4e-29 | 36.63% (243) | putative cell division ATP-binding protein FTSE (septation |
| M. gilvum PYR-GCK | Mflv_3968 | - | 1e-35 | 38.94% (226) | ABC transporter related |
| M. tuberculosis H37Rv | Rv0986 | - | 3e-29 | 36.28% (215) | adhesion component transport ATP-binding protein ABC |
| M. leprae Br4923 | MLBr_00669 | ftsE | 4e-29 | 36.69% (248) | putative cell division ATP-binding protein |
| M. abscessus ATCC 19977 | MAB_3050 | - | 5e-37 | 39.38% (226) | putative glutamate ABC transporter, ATP-binding protein |
| M. marinum M | MMAR_0060 | - | 2e-30 | 37.23% (231) | glutamine ABC transporter, ATP-binding protein |
| M. avium 104 | MAV_4425 | - | 6e-32 | 42.47% (219) | ABC-type transporter, ATPase component |
| M. smegmatis MC2 155 | MSMEG_0647 | phnC | 1e-126 | 87.26% (259) | phosphonate ABC transporter, ATP-binding protein |
| M. thermoresistible (build 8) | TH_4568 | - | 1e-118 | 80.23% (263) | PUTATIVE phosphonate ABC transporter, ATP-binding protein |
| M. ulcerans Agy99 | MUL_0674 | - | 6e-28 | 40.84% (191) | ABC transporter, ATP-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_4425|M.avium_104 --------MT-----------IEMAGVVREYRVGGQPVRALDRISLQLAG
MUL_0674|M.ulcerans_Agy99 --------MTRAQAPTSPRGGVSLHGVDKTFGTAAASVHALRRIDLDIEP
Rv0986|M.tuberculosis_H37Rv --------MNRQP-------IVQLSNLSWTFREGETRRQVLDHITFDFEP
Mb3129c|M.bovis_AF2122/97 --------------------MITLDHVTKQYKSSAR--PALDDINVKIDK
MLBr_00669|M.leprae_Br4923 --------------------MITLDHVSKKYKSLAR--PALDNVNVKIDK
Mvan_2905|M.vanbaalenii_PYR-1 ----MPASRPADHDVAGDDVVIAAHNVTKRFG----NTLALDDVSLEVRR
MSMEG_0647|M.smegmatis_MC2_155 -----------MNPVAGDDVVVIARDVTKRFG----DTLALDHVSLDVHR
TH_4568|M.thermoresistible__bu ------LTAPPVEPVAGDDLVIAVRDATKRFG----GTVALDGVSLDVHR
Mflv_3968|M.gilvum_PYR-GCK ------------------MPMISMKSVNKHFG----PLHVLKDIDLEVGR
MAB_3050|M.abscessus_ATCC_1997 MPVSTDTSTSGESSSAPSVPMIALEHVNKHFG----DLHVLKDITLEIPR
MMAR_0060|M.marinum_M -------MTRQDAGRQP--VCLAARSIHQTLG----GSVVLRGVDLEVPA
: .* : ...
MAV_4425|M.avium_104 GQFVAVVGPSGAGKSTLLHLLGALDSPDSGSILFDGEEIGRLGDEAQSRF
MUL_0674|M.ulcerans_Agy99 GELVAIVGESGSGKTTLLNVIGGIEEANSGSINVAGQDISGCRPRALSKF
Rv0986|M.tuberculosis_H37Rv GEFVALLGQSGSGKSTLLNLISGIEKPTTGDVTINGFAITQKTERDRTLF
Mb3129c|M.bovis_AF2122/97 GEFVFLIGPSGSGKSTFMRLLLAAETPTSGDVRVSKFHVNKLRGRHVPKL
MLBr_00669|M.leprae_Br4923 GEFVFLIGPSGSGKSTFMRLLLGAETPTSGDVRVSKFHVNKLPGRHIPRL
Mvan_2905|M.vanbaalenii_PYR-1 SELLVLLGLSGSGKSTLLRCLNGLHPVTSGTVEVDGTRVDGASKAQLRAL
MSMEG_0647|M.smegmatis_MC2_155 SELLVLLGLSGSGKSTLLRCLNGLHPVTSGTVDVGGTRVDQASGAQLRAL
TH_4568|M.thermoresistible__bu SEFVVLLGLSGSGKSTLLRCLNGLHRVTSGTVEVGGVRVNTASRRELRRL
Mflv_3968|M.gilvum_PYR-GCK GQVVVVLGPSGSGKSTLCRTINRLETIDSGTIAIDGVELP-AEGRKLAQL
MAB_3050|M.abscessus_ATCC_1997 GQVVVILGPSGSGKSTLCRTINRLEPIDGGTIRIDGAILP-EEGRALASL
MMAR_0060|M.marinum_M GTTAAVIGPSGSGKSTLLRTLNRLHEPDSGDILLDGRSVL-RDNP--NDL
. ::* **:**:*: . : . * : . : :
MAV_4425|M.avium_104 RHRRVGFVFQFFNLLPTLSAWENVAIPRLLDGVRLGR--------ARPDA
MUL_0674|M.ulcerans_Agy99 RRDHVGFIFQFFNLIPTLTAKENVEVIVELTG-RGDR--------RR--A
Rv0986|M.tuberculosis_H37Rv RRDQIGIVFQFFNLIPTLTVLENITLPQELAGVSQRK--------AAVVA
Mb3129c|M.bovis_AF2122/97 R-QVIGCVFQDFRLLQQKTVYDNVAFALEVIGKRTDA--------INRVV
MLBr_00669|M.leprae_Br4923 R-QVIGCVFQDFRLLQQKTVYENVAFALEVIGRRSDA--------INQVV
Mvan_2905|M.vanbaalenii_PYR-1 R-RNVGFVFQHFNLVGRLSCLENVLIGGLGQLTFPRYGALTYPKRMRTEA
MSMEG_0647|M.smegmatis_MC2_155 R-RRVGFVFQHFNLVGRLSCLENVLIGGLGRLRLPRYGALTYPRHMRAEA
TH_4568|M.thermoresistible__bu R-RRVGFVFQQFNLVGRLSCLENVLIGGLGRLTLPRYGALTYPRRMRTEA
Mflv_3968|M.gilvum_PYR-GCK R-SDVGMVFQSFNLFAHKTILENVTLAPMKVRKFSKD-------ESREKA
MAB_3050|M.abscessus_ATCC_1997 R-AEVGMVFQSFNLFSHKTILDNVTLAPIKVRKHNKE-------TARKRA
MMAR_0060|M.marinum_M R-QRIGMVFQHFNLFPHRNVVDNVTLAPRKLRGLSNE-------EARMLA
* :* :** *.*. . :*: . .
MAV_4425|M.avium_104 LRLLERVGLGNRTEHRPAELSGGQMQRVAVARALMMDPPLILADEPTGNL
MUL_0674|M.ulcerans_Agy99 TELLDAVELADRAGSFPGQLSGGQQQRVAIARALITNPDLLLADEPMGAL
Rv0986|M.tuberculosis_H37Rv RDLLEKVGMADRERTFPDKLSGGEQQRVAISRALAHNPMLVLADEPTGNL
Mb3129c|M.bovis_AF2122/97 PEVLETVGLSGKANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNL
MLBr_00669|M.leprae_Br4923 PDVLETVGLSGKANRLPDELSGGEQQRVAIARAFVNRPLVLLADEPTGNL
Mvan_2905|M.vanbaalenii_PYR-1 LAHLDRVGLADFAERRADTLSGGQQQRVAIARTLMQAPALLLADEPVASL
MSMEG_0647|M.smegmatis_MC2_155 LAHLDRVGLADYADRRADTLSGGQQQRVAIARTLMQKPALLLADEPVASL
TH_4568|M.thermoresistible__bu LEHLDRVGLAGLAEQRADTLSGGQQQRVAIARTLMQRPSLLLADEPVASL
Mflv_3968|M.gilvum_PYR-GCK MALLERVGVANQADKYPAQLSGGQQQRVAIARSLAMNPKVMLFDEPTSAL
MAB_3050|M.abscessus_ATCC_1997 IELLERVGIASQKDKYPAQLSGGQQQRVAIARSLAMGPKVMLFDEPTSAL
MMAR_0060|M.marinum_M LDQLDRVGLKHKANARPDMLSGGQQQRVAIARALAMAPQVMFFDEATSAL
*: * : . ****: ****::*:: * ::: **. . *
MAV_4425|M.avium_104 DSSTGAAILELLAEVAHEGGGDRLVVMVTHNSDAAAATDR-VITLQDGRV
MUL_0674|M.ulcerans_Agy99 DVATGRRVLELLQQTNRSGLG---VIMVTHNRAAADVADP-IIQMRDGAI
Rv0986|M.tuberculosis_H37Rv DSDTGDKVLDVLLDLTRQAGKT--LIMATHSPSMTQHADR-VVNLQGGRL
Mb3129c|M.bovis_AF2122/97 DPETSRDIMDLLERINRTGTT---VLMATHDHHIVDSMRQRVVELSLGRL
MLBr_00669|M.leprae_Br4923 DPDTSKDIMDLLERINRTGTT---VLMATHDHHIVDSMRQRVVELSLGRL
Mvan_2905|M.vanbaalenii_PYR-1 DPENAGLVMDLLFRVC--IEEKLTVVCTLHQVDLALGWAHRLVGLRNGRK
MSMEG_0647|M.smegmatis_MC2_155 DPENAGVVMDLLFRVC--IEEKLTVVCTLHQVDLALGWAHRLVGLQGGRK
TH_4568|M.thermoresistible__bu DPENAGVVMDLLFQVC--MEQKLTVVCTLHQIDLALGWAHRLVGLRAGRK
Mflv_3968|M.gilvum_PYR-GCK DPEMINEVLAVMTGLA--GDG-MTMLVVTHEMGFARRAANRVVFMSDGAV
MAB_3050|M.abscessus_ATCC_1997 DPEMVNEVLDVMVSLA--KEG-MTMLVVTHEMGFARKAADRIIFMADGAI
MMAR_0060|M.marinum_M DPEMVKGILALIADLG--ADG-MTMVVVTHEMGFARSTSDAVVFMDHGTV
* :: :: :: . *. . :: : *
MAV_4425|M.avium_104 G------SDVLAVAG-------------------
MUL_0674|M.ulcerans_Agy99 VEQRR--NSAPAEASSVRW---------------
Rv0986|M.tuberculosis_H37Rv IPAVNRENQTDQPASTILLPTSYE----------
Mb3129c|M.bovis_AF2122/97 VR-----DEQRGVYGMDR----------------
MLBr_00669|M.leprae_Br4923 VR-----DEWCGIYGMDR----------------
Mvan_2905|M.vanbaalenii_PYR-1 VLDRPAVGLTRDEVMGIYQRVDPAVDSAPRSAVT
MSMEG_0647|M.smegmatis_MC2_155 VLDRPAVGMTRDDVMAVYQRVEPAVTPARRV---
TH_4568|M.thermoresistible__bu VLDRPAVGASHDDVLDIYRRVDPAVIGSPAGR--
Mflv_3968|M.gilvum_PYR-GCK VEDAEPIQFFDNPQTDRAKDFLGKILHH------
MAB_3050|M.abscessus_ATCC_1997 VEDSDPESFFTNPKSERAKDFLGKILGH------
MMAR_0060|M.marinum_M VEAGPPEQLFEAARTERLQRFLAQVL--------