For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRGSYHHGDLKAVILAQAATLVAERGADGVSLRELARAAGVSHAAPAHHFTDRRGVFTALATQGWRMLA AALGESRPEFLDAALAYVRFAVDHPGHYEVMFDRSLVDPDNPELVAAQTEAGAELAAGVGTLDDARARED PQAAGLAAWSLVHGFAMLWLNEAIDTDADAVVTARRVARMLFTPGPVGDAGRG
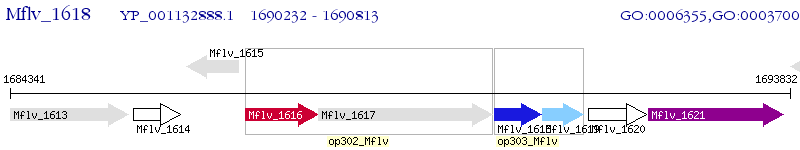
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1618 | - | - | 100% (193) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2110 | - | 1e-08 | 30.49% (164) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2132 | - | 3e-06 | 28.34% (187) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0672c | - | 1e-08 | 27.01% (174) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0653c | - | 1e-08 | 27.01% (174) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_01733 | - | 1e-05 | 30.58% (121) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0499 | - | 4e-47 | 54.14% (181) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4906 | - | 8e-67 | 68.11% (185) | transcriptional regulator |
| M. avium 104 | MAV_0728 | - | 1e-67 | 69.19% (185) | TetR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_5838 | - | 2e-71 | 70.79% (178) | TetR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_1893 | - | 9e-09 | 30.72% (166) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (POSSIBLY |
| M. ulcerans Agy99 | MUL_0500 | - | 2e-66 | 68.11% (185) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5135 | - | 3e-77 | 75.27% (186) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0672c|M.bovis_AF2122/97 --------------------------------------------------
Rv0653c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1618|M.gilvum_PYR-GCK -------------------------------------------------M
Mvan_5135|M.vanbaalenii_PYR-1 ------------------------------------------------MS
MSMEG_5838|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4906|M.marinum_M -------------------------------------------------M
MUL_0500|M.ulcerans_Agy99 -------------------------------------------------M
MAV_0728|M.avium_104 -------------------------------------------------M
MAB_0499|M.abscessus_ATCC_1997 --------------------------------------------------
TH_1893|M.thermoresistible__bu ----------------------------------VNTQMSAGPGGAPGAR
MLBr_01733|M.leprae_Br4923 MTRRSIHTRKPLKWSGCAGQHVRSGKVFDVGRISLSYPSMKPGVKIDARS
Mb0672c|M.bovis_AF2122/97 ---MTSQTGVR-DELLHAGVRLLDDHGPDALQTRKVAAAAGTSTMAVYTH
Rv0653c|M.tuberculosis_H37Rv ---MTSQTGVR-DELLHAGVRLLDDHGPDALQTRKVAAAAGTSTMAVYTH
Mflv_1618|M.gilvum_PYR-GCK TRGSYHHGDLK-AVILAQAATLVAERGADGVSLRELARAAGVSHAAPAHH
Mvan_5135|M.vanbaalenii_PYR-1 SRGTYHHGDLR-ATILARAADLVAERGADGVSLRELARAAGVSHAAPAHH
MSMEG_5838|M.smegmatis_MC2_155 --MAYHHGDLK-AVILQHAATLVAERGADGISLRELARTAGVSHAAPAHH
MMAR_4906|M.marinum_M NRPAYHHGDLR-AVILDEAARLVAQRGADRVSMRELARDAGVSHAAPAHH
MUL_0500|M.ulcerans_Agy99 NRPAYHHGDLR-AVILDEAARLVAQRGADRVSMRELARDAGVSHAAPAHH
MAV_0728|M.avium_104 TRHSYHHGDLP-AVILAEAARLVAERGAERVSLRELARCAGVSHAAPAHH
MAB_0499|M.abscessus_ATCC_1997 --MSYHHGDLR-AAILTSAADMVARRGATELSLRELAREAGVSHAAPAHH
TH_1893|M.thermoresistible__bu TKSPARAGRLSRDTIVNAALTFLDREGWDALTINALATQLGTKGPSLYNH
MLBr_01733|M.leprae_Br4923 ERWREHRKKVR-GEIVDAAFRALDRLGP-EVSVREIAEEAGTAKPKIYRH
: :: . : * : . :* *. *
Mb0672c|M.bovis_AF2122/97 FGGMRGLIAAIAEEGLRQFDVALTVPQTA---DPVADLLAIGTAYRRYAI
Rv0653c|M.tuberculosis_H37Rv FGGMRGLIAAIAEEGLRQFDVALTVPQTA---DPVADLLAIGTAYRRYAI
Mflv_1618|M.gilvum_PYR-GCK FTDRRGVFTALATQGWRMLAAALGESR--------PEFLDAALAYVRFAV
Mvan_5135|M.vanbaalenii_PYR-1 FTDRRGLFTALATEGFRSLAEELSRAR--------PEFLDAALAYVRFAL
MSMEG_5838|M.smegmatis_MC2_155 FTDRRGLFTALATEGYQLLADALKDVR--------GQFIDAAKAYVRFAL
MMAR_4906|M.marinum_M FTDRRGLFTALATQGFQLLAAALVAAR--------GHFADAALAYVRFAI
MUL_0500|M.ulcerans_Agy99 FTDRRGLFTALATQGFQLLAAALVAAR--------GHFADAALAYVRFAI
MAV_0728|M.avium_104 FTDRRGLFTALATQGFELLTRALADAR--------GDFADAALAYVQFAL
MAB_0499|M.abscessus_ATCC_1997 FGDRRGLFTALAADGFTRLADALDGAH--------PDFHAAALAYVEFAL
TH_1893|M.thermoresistible__bu VQSLDDLRRSVRMRVVDDIIGMLNAAGQGR--TRDDAVLAMASAYRSYAH
MLBr_01733|M.leprae_Br4923 FTDKSDLFEAIGIRLRDMLWAAIFPAIDLATDSAREIIRRSVDEYVSLVD
. . .: :: : : . * .
Mb0672c|M.bovis_AF2122/97 ERPHMYRLMFGSTSAHGINVPARDVLTLKVAEIEHQHPSFAHVVRAVHRC
Rv0653c|M.tuberculosis_H37Rv ERPHMYRLMFGSTSAHGINVPARDVLTLKVAEIEHQHPSFAHVVRAVHRC
Mflv_1618|M.gilvum_PYR-GCK DHPGHYEVMFDRSLVD------------------PDNPELVAAQTEAGAE
Mvan_5135|M.vanbaalenii_PYR-1 ANPGHYEVMFDKSLYH------------------ADDPALVDAETAAGSE
MSMEG_5838|M.smegmatis_MC2_155 DHPGHYAVMFDKSLYD------------------DTDPGLVAAASAAGAE
MMAR_4906|M.marinum_M EHPGHYQVMFNRSLHD------------------ADDADLAAAQAAAAAE
MUL_0500|M.ulcerans_Agy99 EHPGHYQVMFNRSLHD------------------ADDADLAAAQAAAAAE
MAV_0728|M.avium_104 DHPGHYQVMFNRSLLD------------------ASDGGLAAAEGAAGAE
MAB_0499|M.abscessus_ATCC_1997 ANPGHYSVMFEPALLD------------------LADGRLAEARDRAAAA
TH_1893|M.thermoresistible__bu HHPGRYSAFTRMPLGG-------------------DDPEFSAAARAAAGP
MLBr_01733|M.leprae_Br4923 KHPNVLRVFIQGRFSAQS---------------EATVRTLDEGREITLTM
.* : : .
Mb0672c|M.bovis_AF2122/97 LLAGRFATALGADDDTAIVATAAQFWSQIHGFVMLELAGFYG-DRGAAVE
Rv0653c|M.tuberculosis_H37Rv LLAGRFATALGADDDTAIVATAAQFWSQIHGFVMLELAGFYG-DRGAAVE
Mflv_1618|M.gilvum_PYR-GCK LAAG-VGTLDDARAREDPQAAGLAAWSLVHGFAMLWLNEAID-TDA---D
Mvan_5135|M.vanbaalenii_PYR-1 LAAG-VGTLDDPRAKDDPQAAALAAWSLVHGFALLWLNGAVN-TTG---D
MSMEG_5838|M.smegmatis_MC2_155 LNRG-VGTLDDPKAKDDPAGAALAAWSMVHGFSLLWLNDAID-TDR---D
MMAR_4906|M.marinum_M LAAG-VATLRDPGARSDPEGAQLAAWSLVHGFSTLWLNDALD-AQVRALD
MUL_0500|M.ulcerans_Agy99 LAAG-VATLRDPGARFDPEGAQLAAWSLVHGFSTLWLNDAFD-AQVRALD
MAV_0728|M.avium_104 LSRG-VATLRDPHARADPGGAELAAWSLVHGFATLWLNDAVD-AAVRAAD
MAB_0499|M.abscessus_ATCC_1997 LDSG-IATLTPQQTSADRITAARAAWSLVHGFVSLWTTGALA-DSRDDAD
TH_1893|M.thermoresistible__bu VIAV-LGSYG--LEGEEAFYAALEFWSALHGFVLLEMTGVMD-DVDSDKV
MLBr_01733|M.leprae_Br4923 AEMFNNELREMELNRAASELAAFAAFGSVASATEWWLGPEPDSPRRMPRE
: :. : .
Mb0672c|M.bovis_AF2122/97 PVLAAMTVNLLVALGDSPERAQCSLRAEQTQKNTLGRAT-----------
Rv0653c|M.tuberculosis_H37Rv PVLAAMTVNLLVALGDSPERAQCSLRAEQTQKNTLGRAT-----------
Mflv_1618|M.gilvum_PYR-GCK AVVTARRVARMLFTPGPVGDAGRG--------------------------
Mvan_5135|M.vanbaalenii_PYR-1 PMATVRRVAGMLFGGEPVTCPT----------------------------
MSMEG_5838|M.smegmatis_MC2_155 PIATVERLGAILFDG-----------------------------------
MMAR_4906|M.marinum_M PMDTVLRIAMMLFKS-----------------------------------
MUL_0500|M.ulcerans_Agy99 PMDTVLRIAMMLFKASRSGCRRFGEGCRRDEGRHHQHHDNPENHRVGVGE
MAV_0728|M.avium_104 PMDTVRRIATMLFAG-----------------------------------
MAB_0499|M.abscessus_ATCC_1997 PLRIASQIAYVLFPEAG---------------------------------
TH_1893|M.thermoresistible__bu FTDMVLRLAAGMEKRS----------------------------------
MLBr_01733|M.leprae_Br4923 QFVAHLTIIMMAVIVGTAETLGISMNPDRPIHDAVPLDRSAS--------
:
Mb0672c|M.bovis_AF2122/97 ----------------
Rv0653c|M.tuberculosis_H37Rv ----------------
Mflv_1618|M.gilvum_PYR-GCK ----------------
Mvan_5135|M.vanbaalenii_PYR-1 ----------------
MSMEG_5838|M.smegmatis_MC2_155 ----------------
MMAR_4906|M.marinum_M ----------------
MUL_0500|M.ulcerans_Agy99 IMHHVVGEVGGEGLGR
MAV_0728|M.avium_104 ----------------
MAB_0499|M.abscessus_ATCC_1997 ----------------
TH_1893|M.thermoresistible__bu ----------------
MLBr_01733|M.leprae_Br4923 ----------------