For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALEPKSVLVDGLRTSYLEAGHGEPVVLLHGGEFGASARIGWEHTIAPLAQRYRVLAPDMLGFGDSAKVVD FTDGRGMRIRHIARFCEVMDVEAAHFVGNSMGAINLLVDVTSDAAVLPVRSLVLICGGGEIQRNEHVDAL YDYDATFEGMRRIVAALFADPAYPTDDEYVRRRYESSIAPGAWESLAAARFRRPGLDPPATPSSARAYDR ITVPTLVLEGRDDKLLPTGWAAEIAGQIRHGRSAVIDDAGHCPQIEQPAAVNQELLDFFEEQT*
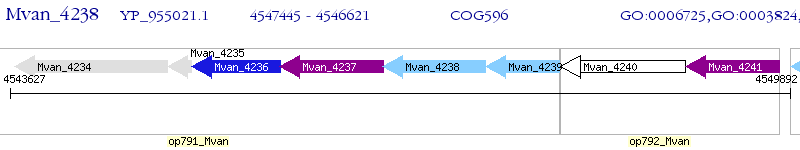
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4238 | - | - | 100% (274) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_0592 | - | 1e-20 | 33.58% (271) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_4414 | - | 3e-18 | 32.95% (264) | alpha/beta hydrolase fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0569 | bpoC | 5e-15 | 27.74% (274) | peroxidase BpoC |
| M. gilvum PYR-GCK | Mflv_2414 | - | 1e-127 | 80.95% (273) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv0554 | bpoC | 5e-15 | 27.74% (274) | peroxidase BpoC |
| M. leprae Br4923 | MLBr_02269 | - | 6e-12 | 28.00% (275) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_4366c | - | 3e-14 | 28.24% (262) | putative hydrolase, alpha/beta fold |
| M. marinum M | MMAR_4771 | - | 1e-111 | 71.21% (264) | hypothetical protein MMAR_4771 |
| M. avium 104 | MAV_0884 | - | 1e-115 | 73.61% (269) | hydrolase, alpha/beta fold family protein, putative |
| M. smegmatis MC2 155 | MSMEG_4860 | - | 1e-116 | 75.65% (271) | hydrolase, alpha/beta fold family protein, putative |
| M. thermoresistible (build 8) | TH_0899 | - | 1e-116 | 74.72% (269) | hydrolase, alpha/beta fold family protein, putative |
| M. ulcerans Agy99 | MUL_0342 | - | 1e-111 | 71.59% (264) | hypothetical protein MUL_0342 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4238|M.vanbaalenii_PYR-1 ----MALEPKSVLVDGLRTSYL-EAGH--GEPVVLLHGGEFGASARIGWE
Mflv_2414|M.gilvum_PYR-GCK --MGVTFERNTVLVEGLTTSYL-EAGQ--GDPIVLLHGGEFGADAALGWE
MSMEG_4860|M.smegmatis_MC2_155 ----MDFERKTIVVDGLTTAYL-EAGDPARDAVVLLHGGEFGVSAELGWE
TH_0899|M.thermoresistible__bu ----VTFDRRTIVVDGLATSYL-EAGS--GDPLVLLHGGEFGASAELGWE
MMAR_4771|M.marinum_M ----MRVHRRAVAVDGLPTSYL-EAGD--GDPVVLLHGGEFGANAEIAWE
MUL_0342|M.ulcerans_Agy99 ----MRIHRRAVAVDGLATSYL-EAGD--GDPVVLLHGGEFGANAEIAWE
MAV_0884|M.avium_104 ----MEIHRKTMSVDGLVTSYL-EAGE--GDPVVLLHGGEFGASAELGWE
Mb0569|M.bovis_AF2122/97 ---------------MINLAYD-DNGT--GDPVVFIAG-RGGAGRTWHPH
Rv0554|M.tuberculosis_H37Rv ---------------MINLAYD-DNGT--GDPVVFIAG-RGGAGRTWHPH
MLBr_02269|M.leprae_Br4923 ---------------MINLAYE-DRGT--GEPVVFIAG-RGGAGRTWQPH
MAB_4366c|M.abscessus_ATCC_199 MTIHTHLQSISTTLGEIAVHDVPAAGGADAPTLVMLHGGGPGASGLSNYE
: * .:*:: * *.. .
Mvan_4238|M.vanbaalenii_PYR-1 HTIAPLAQRYRVLAPDMLGFG----DSAKVVDFTDGRGMRIRHIARFCEV
Mflv_2414|M.gilvum_PYR-GCK ATIDALASRYRVLAPDMLGFG----GSAKVVDFTDGRGMRIRHIARFCDV
MSMEG_4860|M.smegmatis_MC2_155 RTIGALATDHHVLAPDMLGFG----GSAKVVDFTDGRGMRIRHIARFCAQ
TH_0899|M.thermoresistible__bu RNIDALAGYYRVIAPDMLGFG----HSAKVIDFVDGRGMRIRHIARLCAL
MMAR_4771|M.marinum_M HNIGVLGGRFRVLAPDLLGFG----QSAKVVDFVDGRGMRIRHVARFCDV
MUL_0342|M.ulcerans_Agy99 HNIGVLGGRFRVLAPDLLGFG----QSAKVVDFVDGRGMRIRHVARFCDV
MAV_0884|M.avium_104 RNIAALAARYRVLAPDQLGFG----GSAKVIDFVDGRGMRIRHVARFCEL
Mb0569|M.bovis_AF2122/97 QVPAFLAAGYRCITFDNRGIG----ATENAEGFTTQT--MVADTAALIET
Rv0554|M.tuberculosis_H37Rv QVPAFLAAGYRCITFDNRGIG----ATENAEGFTTQT--MVADTAALIET
MLBr_02269|M.leprae_Br4923 QVPAFLAAGYRVITFDNRGIG----ATENTEGFTTQT--MVADTAVLIES
MAB_4366c|M.abscessus_ATCC_199 QNIPALSRTFRILLPDQPGFGGSYRPTGADLDERSITEITVDALFQVLDD
*. .: : * *:* : . : .
Mvan_4238|M.vanbaalenii_PYR-1 MDVEAAHFVGNSMGAINLLVDVTSDAAVLPVRSLVLICGGGEIQR---NE
Mflv_2414|M.gilvum_PYR-GCK MGIAAAHFVGNSMGAINLLVDATSDTPVLPMRSLVAVCGGGDIQR---NE
MSMEG_4860|M.smegmatis_MC2_155 MGVKSAHFVGNSMGAINLLVDTTSDAPLLPVRSLVAICGGGEIQR---NE
TH_0899|M.thermoresistible__bu LGIESAHFVGNSMGAINLLVDATSPAPRLPMRSLVAICGGGEIQR---NE
MMAR_4771|M.marinum_M LGVDSAHFVGNSMGAIMLLTDTTSDAPMLPVQTMALICGGGQIQQ---NS
MUL_0342|M.ulcerans_Agy99 LGVGSAHFVGNSMGAIMLLTDTTSDAPMLPVRTMALICGGGQIQQ---NS
MAV_0884|M.avium_104 LGVESAHFVGNSMGAINLLTDATSDSPLWPVRSAVIICGGGEIQQ---NE
Mb0569|M.bovis_AF2122/97 LDIAPARVVGVSMGAFIAQELMVVAPELVSSAVLMATRGRLDRARQFFNK
Rv0554|M.tuberculosis_H37Rv LDIAPARVVGVSMGAFIAQELMVVAPELVSSAVLMATRGRLDRARQFFNK
MLBr_02269|M.leprae_Br4923 LGAVPARIVGVSMGSFIAQELMVARPELVRAAVLMATRGRLDRTRQFFHA
MAB_4366c|M.abscessus_ATCC_199 LAVGSFHLLGNSLGGAAAIRMAQLRPERVTRLVLMAPGGGWLPFGPTPTE
: . :.:* *:*. . *
Mvan_4238|M.vanbaalenii_PYR-1 HVDALYDYDATFEGMRRIVAALFADPAYP---TDDEYVRRRYESSIAPGA
Mflv_2414|M.gilvum_PYR-GCK HVSALYDYDATLEGMRRIVTALFADPSYP---ADDEYVRRRYESSIAPGA
MSMEG_4860|M.smegmatis_MC2_155 HMAALYDYDATLEGMRRIVAALFADPSYP---ADEDYVRRRYESSIAPGA
TH_0899|M.thermoresistible__bu HMAALYDYDATEPAMRRIVTALFFDPAFP---ADDAYVRRRYESSILPGA
MMAR_4771|M.marinum_M HFDALQSYDATLPAMRRIVAALFHDARYP---ADMDYVQRRYDSSIAPGA
MUL_0342|M.ulcerans_Agy99 HFDALQSYDATLPAMRRIVAALFHDARYP---ADMDYVQRRYDSSIAPGA
MAV_0884|M.avium_104 YFEALQNYDATLPAMRRIVEALFFDPAYP---ADDDYVRRRHESSTAPGA
Mb0569|M.bovis_AF2122/97 AEAELYDSGVQLPPTYDARARLLENFSRK---TLNDDVAVGDWIAMFS-M
Rv0554|M.tuberculosis_H37Rv AEAELYDSGVQLPPTYDARARLLENFSRK---TLNDDVAVGDWIAMFS-M
MLBr_02269|M.leprae_Br4923 AEAEFHDSGIQLPSGYNAKVRLLENLSRK---TLNDDVAVADWIAMFN-M
MAB_4366c|M.abscessus_ATCC_199 GQKEMFRYFNGEGPTEKKMAAFIRAMVFDHKQFGRDVVSRRYQASLDEGH
: :: * :
Mvan_4238|M.vanbaalenii_PYR-1 WESLAAARFRRPGLDPPATPSSARAYDRITVPTLVLEGRDDKLLPTGWAA
Mflv_2414|M.gilvum_PYR-GCK WESLAAARFRRPGLEPPPTPSSSRAYERITVPTLVVEGGADKLLPRGWAA
MSMEG_4860|M.smegmatis_MC2_155 WEALAAARFRRPGLEPPAAPSSARAYERIGVPVLIVEGERDKLLPSGWAA
TH_0899|M.thermoresistible__bu WETLAAARFRRPGLEPPAAPSAARAYHRIGVPTLIVEGAGDKLLPPGWAA
MMAR_4771|M.marinum_M WEAVAAARFRRPGAPPSATPSSSRRYDRITVPTMLVEGRNDKLLPSGWGE
MUL_0342|M.ulcerans_Agy99 WEAVAAARFRRPGAPPSATPSSSRRYDRITVPTMLVEGRNDKLLPSGWGE
MAV_0884|M.avium_104 WEAIAAARFRRPGATPSPTPSSARAYERIGVPTLVVEGRGDKLLPAGWSA
Mb0569|M.bovis_AF2122/97 WPIKSTPGLR-CQLDCAPQTNRLPAYRNIAAPVLVIGFADDVVTPPYLGR
Rv0554|M.tuberculosis_H37Rv WPIKSTPGLR-CQLDCAPQTNRLPAYRNIAAPVLVIGFADDVVTPPYLGR
MLBr_02269|M.leprae_Br4923 WPIKSTPGLR-CQTDVAPQNNRLPAYRSIAAPVLVIGFAEDVVTPPSLGR
MAB_4366c|M.abscessus_ATCC_199 IEFYHHYNAAFAKRN--GMDPLWRDLHTITAPTLLLWGRDDRTITLEGAQ
* .*.::: * . .
Mvan_4238|M.vanbaalenii_PYR-1 EIAGQIRHGRSAVIDDAGHCPQIEQPAAVNQELLDFFEEQT---------
Mflv_2414|M.gilvum_PYR-GCK EIAGQIAGARSAVIDEAGHCPQIEQPEAVIGLLLTFFEEQR---------
MSMEG_4860|M.smegmatis_MC2_155 QIAAQISGATSTVVADAGHCPQIEQPDTVNRLLSDFLKEVS---------
TH_0899|M.thermoresistible__bu DIAAQIPDARSAVIADAGHCPQIEQPAAVNELLLDFLTTVNTRDREVATR
MMAR_4771|M.marinum_M QIAGQIRNGHSQVVDRAGHCPQIEQAGTVNELMLDFLTR-----------
MUL_0342|M.ulcerans_Agy99 QIAGQIRNGHSQVVDRAGHCPQIEQAGTVNELMLDFLSR-----------
MAV_0884|M.avium_104 QIAKQIDGARSVVVDDAGHCPQIEQPSVVNQLLMDFLATA----------
Mb0569|M.bovis_AF2122/97 EVADALPNGRYLQIPDAGHLGFFERPEAVNTAMLKFFASVKA--------
Rv0554|M.tuberculosis_H37Rv EVADALPNGRYLQIPDAGHLGFFERPEAVNTAMLKFFASVKA--------
MLBr_02269|M.leprae_Br4923 EVADVLPNGRYLQIPDAGHLGFFERPEAVNAAALQFFAGVKGEWP-----
MAB_4366c|M.abscessus_ATCC_199 MMLKHISDVQLHVFGRCGHWVQLERQAEFDRLVADFLGEPA---------
: : . .** :*: . *: