For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVHRRAVAVDGLPTSYLEAGDGDPVVLLHGGEFGANAEIAWEHNIGVLGGRFRVLAPDLLGFGQSAKVVD FVDGRGMRIRHVARFCDVLGVDSAHFVGNSMGAIMLLTDTTSDAPMLPVQTMALICGGGQIQQNSHFDAL QSYDATLPAMRRIVAALFHDARYPADMDYVQRRYDSSIAPGAWEAVAAARFRRPGAPPSATPSSSRRYDR ITVPTMLVEGRNDKLLPSGWGEQIAGQIRNGHSQVVDRAGHCPQIEQAGTVNELMLDFLTR*
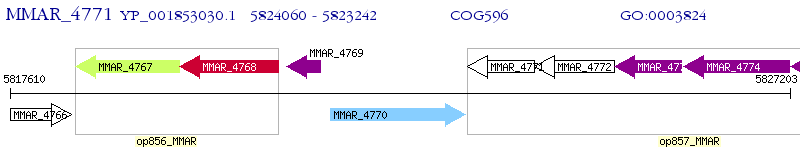
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4771 | - | - | 100% (272) | hypothetical protein MMAR_4771 |
| M. marinum M | MMAR_1502 | - | 9e-26 | 31.65% (278) | hypothetical protein MMAR_1502 |
| M. marinum M | MMAR_5064 | bphD | 2e-12 | 27.86% (280) | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3600c | bphD | 1e-11 | 27.34% (278) | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD |
| M. gilvum PYR-GCK | Mflv_2414 | - | 1e-105 | 67.29% (269) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3569c | bphD | 1e-11 | 27.34% (278) | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD |
| M. leprae Br4923 | MLBr_00685 | - | 8e-06 | 23.33% (300) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_4366c | - | 2e-16 | 27.70% (278) | putative hydrolase, alpha/beta fold |
| M. avium 104 | MAV_0884 | - | 1e-116 | 72.59% (270) | hydrolase, alpha/beta fold family protein, putative |
| M. smegmatis MC2 155 | MSMEG_4860 | - | 1e-104 | 68.75% (272) | hydrolase, alpha/beta fold family protein, putative |
| M. thermoresistible (build 8) | TH_0899 | - | 1e-106 | 67.16% (271) | hydrolase, alpha/beta fold family protein, putative |
| M. ulcerans Agy99 | MUL_0342 | - | 1e-155 | 98.16% (272) | hypothetical protein MUL_0342 |
| M. vanbaalenii PYR-1 | Mvan_4238 | - | 1e-111 | 71.21% (264) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3600c|M.bovis_AF2122/97 MTATEELTFESTSRFAEVDVDGPLKLHYHEAGVGNDQTVVLLHGGGPGAA
Rv3569c|M.tuberculosis_H37Rv MTATEELTFESTSRFAEVDVDGPLKLHYHEAGVGNDQTVVLLHGGGPGAA
MAB_4366c|M.abscessus_ATCC_199 MTIHTHLQSISTT-LGEIAVH-----DVPAAGGADAPTLVMLHGGGPGAS
MMAR_4771|M.marinum_M ------MRVHRRA----VAVDG-LPTSYLEAGD--GDPVVLLHGGEFGAN
MUL_0342|M.ulcerans_Agy99 ------MRIHRRA----VAVDG-LATSYLEAGD--GDPVVLLHGGEFGAN
MSMEG_4860|M.smegmatis_MC2_155 ------MDFERKT----IVVDG-LTTAYLEAGDPARDAVVLLHGGEFGVS
TH_0899|M.thermoresistible__bu ------VTFDRRT----IVVDG-LATSYLEAGS--GDPLVLLHGGEFGAS
Mflv_2414|M.gilvum_PYR-GCK ----MGVTFERNT----VLVEG-LTTSYLEAGQ--GDPIVLLHGGEFGAD
Mvan_4238|M.vanbaalenii_PYR-1 ------MALEPKS----VLVDG-LRTSYLEAGH--GEPVVLLHGGEFGAS
MAV_0884|M.avium_104 ------MEIHRKT----MSVDG-LVTSYLEAGE--GDPVVLLHGGEFGAS
MLBr_00685|M.leprae_Br4923 MPACQHQRHRPVVACGTRGTTFRVGSMRQPIHLGSGEPVLLLHPFMMSQT
. .:::** .
Mb3600c|M.bovis_AF2122/97 SWTNFSRNIAVLARHFHVLAVDQPGYGHSDK----RAEHGQFNRYAAMAL
Rv3569c|M.tuberculosis_H37Rv SWTNFSRNIAVLARHFHVLAVDQPGYGHSDK----RAEHGQFNRYAAMAL
MAB_4366c|M.abscessus_ATCC_199 GLSNYEQNIPALSRTFRILLPDQPGFGGSYRPTGADLDERSITEITVDAL
MMAR_4771|M.marinum_M AEIAWEHNIGVLGGRFRVLAPDLLGFGQSAK----VVDFVDGRGMRIRHV
MUL_0342|M.ulcerans_Agy99 AEIAWEHNIGVLGGRFRVLAPDLLGFGQSAK----VVDFVDGRGMRIRHV
MSMEG_4860|M.smegmatis_MC2_155 AELGWERTIGALATDHHVLAPDMLGFGGSAK----VVDFTDGRGMRIRHI
TH_0899|M.thermoresistible__bu AELGWERNIDALAGYYRVIAPDMLGFGHSAK----VIDFVDGRGMRIRHI
Mflv_2414|M.gilvum_PYR-GCK AALGWEATIDALASRYRVLAPDMLGFGGSAK----VVDFTDGRGMRIRHI
Mvan_4238|M.vanbaalenii_PYR-1 ARIGWEHTIAPLAQRYRVLAPDMLGFGDSAK----VVDFTDGRGMRIRHI
MAV_0884|M.avium_104 AELGWERNIAALAARYRVLAPDQLGFGGSAK----VIDFVDGRGMRIRHV
MLBr_00685|M.leprae_Br4923 VWEIVAPQLANTER-YEVFAPTMAAHHGGPS----ASSWMMSVSTLADHV
: ..:: .. . . :
Mb3600c|M.bovis_AF2122/97 KGLFDQLGLGRVPLVGNSLGGG-TAVRFALDYPARAGRLVLMGPGGLSIN
Rv3569c|M.tuberculosis_H37Rv KGLFDQLGLGRVPLVGNSLGGG-TAVRFALDYPARAGRLVLMGPGGLSIN
MAB_4366c|M.abscessus_ATCC_199 FQVLDDLAVGSFHLLGNSLGGA-AAIRMAQLRPERVTRLVLMAPGGGWL-
MMAR_4771|M.marinum_M ARFCDVLGVDSAHFVGNSMGAIMLLTDTTSDAPMLPVQTMALICGGGQI-
MUL_0342|M.ulcerans_Agy99 ARFCDVLGVGSAHFVGNSMGAIMLLTDTTSDAPMLPVRTMALICGGGQI-
MSMEG_4860|M.smegmatis_MC2_155 ARFCAQMGVKSAHFVGNSMGAINLLVDTTSDAPLLPVRSLVAICGGGEI-
TH_0899|M.thermoresistible__bu ARLCALLGIESAHFVGNSMGAINLLVDATSPAPRLPMRSLVAICGGGEI-
Mflv_2414|M.gilvum_PYR-GCK ARFCDVMGIAAAHFVGNSMGAINLLVDATSDTPVLPMRSLVAVCGGGDI-
Mvan_4238|M.vanbaalenii_PYR-1 ARFCEVMDVEAAHFVGNSMGAINLLVDVTSDAAVLPVRSLVLICGGGEI-
MAV_0884|M.avium_104 ARFCELLGVESAHFVGNSMGAINLLTDATSDSPLWPVRSAVIICGGGEI-
MLBr_00685|M.leprae_Br4923 EQQLDELGWDTAHLVGNSLGGWVVFELERRGRARSVTG--IAPAGGWTR-
: ::***:*. . **
Mb3600c|M.bovis_AF2122/97 LFAPDPTEGVKRLSKFS--VAPTRENLEAFLRVMVYDKNLITPELVDQRF
Rv3569c|M.tuberculosis_H37Rv LFAPDPTEGVKRLSKFS--VAPTRENLEAFLRVMVYDKNLITPELVDQRF
MAB_4366c|M.abscessus_ATCC_199 PFGPTPTEGQKEMFRYFNGEGPTEKKMAAFIRAMVFDHKQFGRDVVSRRY
MMAR_4771|M.marinum_M ----QQNSHFDALQSYDATLPAMRRIVAALFHDARYP---ADMDYVQRRY
MUL_0342|M.ulcerans_Agy99 ----QQNSHFDALQSYDATLPAMRRIVAALFHDARYP---ADMDYVQRRY
MSMEG_4860|M.smegmatis_MC2_155 ----QRNEHMAALYDYDATLEGMRRIVAALFADPSYP---ADEDYVRRRY
TH_0899|M.thermoresistible__bu ----QRNEHMAALYDYDATEPAMRRIVTALFFDPAFP---ADDAYVRRRY
Mflv_2414|M.gilvum_PYR-GCK ----QRNEHVSALYDYDATLEGMRRIVTALFADPSYP---ADDEYVRRRY
Mvan_4238|M.vanbaalenii_PYR-1 ----QRNEHVDALYDYDATFEGMRRIVAALFADPAYP---TDDEYVRRRY
MAV_0884|M.avium_104 ----QQNEYFEALQNYDATLPAMRRIVEALFFDPAYP---ADDDYVRRRH
MLBr_00685|M.leprae_Br4923 ----WSLAKFEVIAKFVAGMPLVAVARGLGSRALSLPFSHRLASLPLTGS
: :
Mb3600c|M.bovis_AF2122/97 ALASTPESLTATRAMG----KSFAGADFEAGMMWREVYR-LRQPVLLIWG
Rv3569c|M.tuberculosis_H37Rv ALASTPESLTATRAMG----KSFAGADFEAGMMWREVYR-LRQPVLLIWG
MAB_4366c|M.abscessus_ATCC_199 QASLDEGHIEFYHHYN----AAFAKRN-GMDPLWRDLHT-ITAPTLLLWG
MMAR_4771|M.marinum_M DSSIAPGAWEAVAAAR----FRRPGAPPSATPSSSRRYDRITVPTMLVEG
MUL_0342|M.ulcerans_Agy99 DSSIAPGAWEAVAAAR----FRRPGAPPSATPSSSRRYDRITVPTMLVEG
MSMEG_4860|M.smegmatis_MC2_155 ESSIAPGAWEALAAAR----FRRPGLEPPAAPSSARAYERIGVPVLIVEG
TH_0899|M.thermoresistible__bu ESSILPGAWETLAAAR----FRRPGLEPPAAPSAARAYHRIGVPTLIVEG
Mflv_2414|M.gilvum_PYR-GCK ESSIAPGAWESLAAAR----FRRPGLEPPPTPSSSRAYERITVPTLVVEG
Mvan_4238|M.vanbaalenii_PYR-1 ESSIAPGAWESLAAAR----FRRPGLDPPATPSSARAYDRITVPTLVLEG
MAV_0884|M.avium_104 ESSTAPGAWEAIAAAR----FRRPGATPSPTPSSARAYERIGVPTLVVEG
MLBr_00685|M.leprae_Br4923 PAGLSEHQLRNIVEDAGHCPAYFLLLIKTLLQPGLRELADITVPVLLVLC
. : *.:::
Mb3600c|M.bovis_AF2122/97 REDRVNP-LDGALVALKTIPR-AQLHVFGQCGHWVQVEKFDEFNKLTIEF
Rv3569c|M.tuberculosis_H37Rv REDRVNP-LDGALVALKTIPR-AQLHVFGQCGHWVQVEKFDEFNKLTIEF
MAB_4366c|M.abscessus_ATCC_199 RDDRTIT-LEGAQMMLKHISD-VQLHVFGRCGHWVQLERQAEFDRLVADF
MMAR_4771|M.marinum_M RNDKLLP-SGWGEQIAGQIRN-GHSQVVDRAGHCPQIEQAGTVNELMLDF
MUL_0342|M.ulcerans_Agy99 RNDKLLP-SGWGEQIAGQIRN-GHSQVVDRAGHCPQIEQAGTVNELMLDF
MSMEG_4860|M.smegmatis_MC2_155 ERDKLLP-SGWAAQIAAQISG-ATSTVVADAGHCPQIEQPDTVNRLLSDF
TH_0899|M.thermoresistible__bu AGDKLLP-PGWAADIAAQIPD-ARSAVIADAGHCPQIEQPAAVNELLLDF
Mflv_2414|M.gilvum_PYR-GCK GADKLLP-RGWAAEIAGQIAG-ARSAVIDEAGHCPQIEQPEAVIGLLLTF
Mvan_4238|M.vanbaalenii_PYR-1 RDDKLLP-TGWAAEIAGQIRH-GRSAVIDDAGHCPQIEQPAAVNQELLDF
MAV_0884|M.avium_104 RGDKLLP-AGWSAQIAKQIDG-ARSVVVDDAGHCPQIEQPSVVNQLLMDF
MLBr_00685|M.leprae_Br4923 EKDRVLPPPKYSRHFTDHLPERTQVTKFDGMGHVPMFEAPEVITKLITNF
*: . . : . ** .* . *
Mb3600c|M.bovis_AF2122/97 LGGGR----------
Rv3569c|M.tuberculosis_H37Rv LGGGR----------
MAB_4366c|M.abscessus_ATCC_199 LGEPA----------
MMAR_4771|M.marinum_M LTR------------
MUL_0342|M.ulcerans_Agy99 LSR------------
MSMEG_4860|M.smegmatis_MC2_155 LKEVS----------
TH_0899|M.thermoresistible__bu LTTVNTRDREVATR-
Mflv_2414|M.gilvum_PYR-GCK FEEQR----------
Mvan_4238|M.vanbaalenii_PYR-1 FEEQT----------
MAV_0884|M.avium_104 LATA-----------
MLBr_00685|M.leprae_Br4923 INQCSKPAEAASPAS
: