For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRSDSEYRVRVGTAEISVTDTGGDGAPVVLLHGGGPGASGISNYSRNIGALAASYRVIVPDMPGYGRSTK YVDHSDPFGYLADSIRGMLDEMGVQQAHLVGNSYGGAAALRLALDTPHRADKLVLMGPGGIGTTRGLPTP GLKSLLGYYAGEGPSREKLATFIRSYLVYDGPSVPDELIDLRYQASLDPQVVADPPLRRPSGLRTLWRMD LTRDKRLRRLPNPTLVLWGRDDKVNRPAGGPMLLNMMPNAELVMTSHTGHWMQWERAELFNELVSDFLSP DTRLASS*
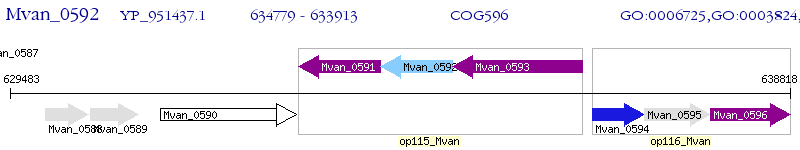
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0592 | - | - | 100% (288) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_5307 | - | 2e-45 | 40.15% (259) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_4414 | - | 1e-35 | 34.23% (260) | alpha/beta hydrolase fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3600c | bphD | 4e-46 | 39.15% (258) | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD |
| M. gilvum PYR-GCK | Mflv_1464 | - | 1e-46 | 40.00% (265) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3569c | bphD | 4e-46 | 39.15% (258) | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD |
| M. leprae Br4923 | MLBr_00685 | - | 2e-05 | 33.04% (115) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_4366c | - | 7e-50 | 39.93% (278) | putative hydrolase, alpha/beta fold |
| M. marinum M | MMAR_1502 | - | 1e-124 | 78.03% (264) | hypothetical protein MMAR_1502 |
| M. avium 104 | MAV_2517 | - | 1e-123 | 80.31% (259) | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase |
| M. smegmatis MC2 155 | MSMEG_6037 | - | 2e-45 | 39.23% (260) | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase |
| M. thermoresistible (build 8) | TH_1741 | - | 8e-48 | 40.30% (268) | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD |
| M. ulcerans Agy99 | MUL_4140 | bphD | 1e-46 | 39.77% (264) | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1502|M.marinum_M ----------------MTTTTLAERTVTVDGKPIFFAEAG--SGPTVVLL
MAV_2517|M.avium_104 ----------------MTTSNMADHAVTVNGKSIFFAEKG--TGAPVLLL
Mvan_0592|M.vanbaalenii_PYR-1 ----------------MTRSD-SEYRVRVGTAEISVTDTGG-DGAPVVLL
Mflv_1464|M.gilvum_PYR-GCK MTSFAAEAAQQQEITFESTSRYAQVR---EDMRLHYHEAGVGNPKTVVLL
TH_1741|M.thermoresistible__bu MTSYIQEAENQVEITFESTSRYAEVKAGDRDMRLHYHEAGIGHAQKVVLL
MSMEG_6037|M.smegmatis_MC2_155 ------MVTATQEITFESTSRFADVQAGELAMRLHYHEAGDPSAQTIVLL
Mb3600c|M.bovis_AF2122/97 -------MTATEELTFESTSRFAEVD-VDGPLKLHYHEAGVGNDQTVVLL
Rv3569c|M.tuberculosis_H37Rv -------MTATEELTFESTSRFAEVD-VDGPLKLHYHEAGVGNDQTVVLL
MUL_4140|M.ulcerans_Agy99 ---MTVTQSKTAEHTFESTSRYAEVD-VDGPLKLHYHEAGVGNDQTVVLL
MAB_4366c|M.abscessus_ATCC_199 -------MTIHTHLQSISTT-LGEIA------VHDVPAAGGADAPTLVML
MLBr_00685|M.leprae_Br4923 --------MPACQHQRHRPVVACGTRGTTFRVGSMRQPIHLGSGEPVLLL
:::*
MMAR_1502|M.marinum_M HGGGPGATGLSNYARNIDLLAQRFRLIIPDMPGYGRSSKGV----DQSDP
MAV_2517|M.avium_104 HGGGPGASGVSNYSRNIDPLAEKFRVIVPDMPGYGRSAKGV----DQSDP
Mvan_0592|M.vanbaalenii_PYR-1 HGGGPGASGISNYSRNIGALAASYRVIVPDMPGYGRSTKYV----DHSDP
Mflv_1464|M.gilvum_PYR-GCK HGGGPGASSWSNFGRNIPVLAKHFHVLAVDQPGYGHSDKHT----EHEQY
TH_1741|M.thermoresistible__bu HGGGPGASSWSNFGRNIAVLARHFHVIAVDQPGYGHSDKHT----EHEQY
MSMEG_6037|M.smegmatis_MC2_155 HGGGPGAASWSNFGRNIAVLAEHFHVLAVDQPGYGLSDKHT----EHEQY
Mb3600c|M.bovis_AF2122/97 HGGGPGAASWTNFSRNIAVLARHFHVLAVDQPGYGHSDKRA----EHGQF
Rv3569c|M.tuberculosis_H37Rv HGGGPGAASWTNFSRNIAVLARHFHVLAVDQPGYGHSDKRA----EHGQF
MUL_4140|M.ulcerans_Agy99 HGGGPGASSWSNFARNIEVLAQQFHVLAVDQPGYGHSDKRA----EHGQF
MAB_4366c|M.abscessus_ATCC_199 HGGGPGASGLSNYEQNIPALSRTFRILLPDQPGFGGSYRPTGADLDERSI
MLBr_00685|M.leprae_Br4923 HPFMMSQTVWEIVAPQLAN-TERYEVFAPTMAAHHGGPSAS----SWMMS
* . : :: : :.:: ... . .
MMAR_1502|M.marinum_M FGYLASAIRGLLDELRIP-SAHLVGNSYGGAAALRLALDSPHRVDRLVLM
MAV_2517|M.avium_104 FGYLADTMRGLLDELGID-SAHLVGNSYGGSCALRLALDNPRRVGKLVLM
Mvan_0592|M.vanbaalenii_PYR-1 FGYLADSIRGMLDEMGVQ-QAHLVGNSYGGAAALRLALDTPHRADKLVLM
Mflv_1464|M.gilvum_PYR-GCK NRYSATALLNLFDHLGIE-KAALVGNSLGGGTAVRFALDNGKRAGKLVLM
TH_1741|M.thermoresistible__bu NRYSATALLALFDHLGIE-RADLVGNSLGGGTAVRFALDHPKRAGRLVLM
MSMEG_6037|M.smegmatis_MC2_155 NRYSAKAVRGLLDHLGVEGRVPLLGNSLGGGTAVRFALDYPDRAGKLVLM
Mb3600c|M.bovis_AF2122/97 NRYAAMALKGLFDQLGLG-RVPLVGNSLGGGTAVRFALDYPARAGRLVLM
Rv3569c|M.tuberculosis_H37Rv NRYAAMALKGLFDQLGLG-RVPLVGNSLGGGTAVRFALDYPARAGRLVLM
MUL_4140|M.ulcerans_Agy99 NHYAARALKELFDQLGLG-RVLLVGNSLGGGTAVRFALDYPDRAGRLVLM
MAB_4366c|M.abscessus_ATCC_199 TEITVDALFQVLDDLAVG-SFHLLGNSLGGAAAIRMAQLRPERVTRLVLM
MLBr_00685|M.leprae_Br4923 VSTLADHVEQQLDELGWD-TAHLVGNSLGG--WVVFELERRGRARSVTGI
. : :*.: *:*** ** : : *. :. :
MMAR_1502|M.marinum_M GPGGIGTTRSAP--TAGLKTLLSYYAGDGPSRAKLAHLIRTYLVYEGDSV
MAV_2517|M.avium_104 GPGGIGTTRGLP--TAGLNSLLAYYGGRGPSRDKLEAFIRNYLVYDGASV
Mvan_0592|M.vanbaalenii_PYR-1 GPGGIGTTRGLP--TPGLKSLLGYYAGEGPSREKLATFIRSYLVYDGPSV
Mflv_1464|M.gilvum_PYR-GCK GPGGLSVNLFSPDPTEGVKLLGRFA--AEPTRENIEKFLR-IMVFDQAMI
TH_1741|M.thermoresistible__bu GPGGLSVNMFAPDPTEGVKLLARFN--AEPTRENLEAFLR-IMVYDQSLI
MSMEG_6037|M.smegmatis_MC2_155 GPGGLSVNLFAPDPTEGVKALAKFN--VEPTRENLEAFLR-IMVFDQKLI
Mb3600c|M.bovis_AF2122/97 GPGGLSINLFAPDPTEGVKRLSKFS--VAPTRENLEAFLR-VMVYDKNLI
Rv3569c|M.tuberculosis_H37Rv GPGGLSINLFAPDPTEGVKRLSKFS--VAPTRENLEAFLR-VMVYDKNLI
MUL_4140|M.ulcerans_Agy99 GPGGLSINLFAPDPTEGVKRLGKFS--VAPTRENLEAFLR-VMVYDQKLI
MAB_4366c|M.abscessus_ATCC_199 APGGGWL-PFGPTPTEGQKEMFRYFNGEGPTEKKMAAFIR-AMVFDHKQF
MLBr_00685|M.leprae_Br4923 APAGGWTRWSLAKFEVIAKFVAGMP-LVAVARGLGSRALS--LPFSHRLA
.*.* . : : :. : : :.
MMAR_1502|M.marinum_M PDELIDLRYQASIDPAVIADPPLRRPNG---LRTLWRMDLTRDRRLRQLP
MAV_2517|M.avium_104 PDELIDLRYQASVDPEVVADPPLRRPSGPMALRTLWRMDLARDRRLKRLR
Mvan_0592|M.vanbaalenii_PYR-1 PDELIDLRYQASLDPQVVADPPLRRPSG---LRTLWRMDLTRDKRLRRLP
Mflv_1464|M.gilvum_PYR-GCK TPELVEERFQIASTPESLAAT---KAMGKSFAGADFELGMMWR-DVYKLR
TH_1741|M.thermoresistible__bu TEELIDERFAIAATPESLAAT---RAMGKSFMGPDFELGMMWR-DAYKLR
MSMEG_6037|M.smegmatis_MC2_155 TPELVEERFAIASTPESLAAT---RAMGKSFAGADFELGMMWR-EVYKLR
Mb3600c|M.bovis_AF2122/97 TPELVDQRFALASTPESLTAT---RAMGKSFAGADFEAGMMWR-EVYRLR
Rv3569c|M.tuberculosis_H37Rv TPELVDQRFALASTPESLTAT---RAMGKSFAGADFEAGMMWR-EVYRLR
MUL_4140|M.ulcerans_Agy99 TPELVDQRFELARTPESLAAT---RAMGKSFAAADFELGMMWR-EVYKLR
MAB_4366c|M.abscessus_ATCC_199 GRDVVSRRYQASLDEGHIEFY---HHYNAAFAKRN-GMDPLWR-DLHTIT
MLBr_00685|M.leprae_Br4923 SLPLTGSPAGLSEHQLRNIVEDAGHCPAYFLLLIKTLLQPGLR-ELADIT
: : : :
MMAR_1502|M.marinum_M TPTLVLWGRDDKIN-RPAGGPKLLNLMPN-AELVMTSRTGHWMQWERAEL
MAV_2517|M.avium_104 TPTLVLWGRDDKVN-RPAGGPMLLNLMPN-AELVMTSRTGHWVQWERADL
Mvan_0592|M.vanbaalenii_PYR-1 NPTLVLWGRDDKVN-RPAGGPMLLNMMPN-AELVMTSHTGHWMQWERAEL
Mflv_1464|M.gilvum_PYR-GCK QRVLLIWGREDRVN-PLDGALVALKQIPR-VQLHVFGQCGHWAQLEKFDE
TH_1741|M.thermoresistible__bu QRVLLIWGREDRVN-PLDGALVALKQIPR-VQLHVFGQCGHWAQVEKFDE
MSMEG_6037|M.smegmatis_MC2_155 QPVLLIWGREDRVN-PLDGALVAVKQIPR-VQLHVFGQCGHWAQVEKFDE
Mb3600c|M.bovis_AF2122/97 QPVLLIWGREDRVN-PLDGALVALKTIPR-AQLHVFGQCGHWVQVEKFDE
Rv3569c|M.tuberculosis_H37Rv QPVLLIWGREDRVN-PLDGALVALKTIPR-AQLHVFGQCGHWVQVEKFDE
MUL_4140|M.ulcerans_Agy99 QPVLLIWGREDRVN-PLDGALVALKTIPR-AQLHVFGQCGHWAQVEKFDE
MAB_4366c|M.abscessus_ATCC_199 APTLLLWGRDDRTI-TLEGAQMMLKHISD-VQLHVFGRCGHWVQLERQAE
MLBr_00685|M.leprae_Br4923 VPVLLVLCEKDRVLPPPKYSRHFTDHLPERTQVTKFDGMGHVPMFEAPEV
.*:: ..*: . . :. .:: . ** *
MMAR_1502|M.marinum_M FNQLVAEFLSQSSAFAS------
MAV_2517|M.avium_104 FNKLVTEFLHNTSVFAQ------
Mvan_0592|M.vanbaalenii_PYR-1 FNELVSDFLSPDTRLASS-----
Mflv_1464|M.gilvum_PYR-GCK FNKLTIDFLTN------------
TH_1741|M.thermoresistible__bu FNRLTIDFLEDGAKEAKS-----
MSMEG_6037|M.smegmatis_MC2_155 FNKLTIDFLGG------------
Mb3600c|M.bovis_AF2122/97 FNKLTIEFLGGGR----------
Rv3569c|M.tuberculosis_H37Rv FNKLTIEFLGGGR----------
MUL_4140|M.ulcerans_Agy99 FNRLTIDFLGGAR----------
MAB_4366c|M.abscessus_ATCC_199 FDRLVADFLGEPA----------
MLBr_00685|M.leprae_Br4923 ITKLITNFINQCSKPAEAASPAS
: .* :*: