For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGNAVRLQAINNVEAYVPPAISFDPTEAPGQIFGSNVFTKAEMQLRLPKSVYKSVVATIEKGAKLDPAIA DAVASAMKDWALSKGATHYAHVFYPMTGLTAEKHDSFLEPISDGETLAEFAGKTLIQGEPDASSFPSGGL RSTFEARGYTGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDYKTPLLRSQQAMGVHAERILALFGHD NLQKVVSFCGPEQEYFLVDRHFFLARPDLVNAGRTLFGAKPPKGQEFDDHYFGAVPERVLGFMMDTEREL FKLGIPAKTRHNEVAPAQFEVAPMFERANIASDHQQLLMTVFKTVAKKHGMECLFHEKPFAGVNGSGKHV NFSVGNADLGSLLVPGDTPHENAQFLVFCAAIIRAVHKFGGLLRVSVASATNDHRLGANEAPPAIISIFL GEQLADVFDQIAKGAATSSKGKGVMHIGVDTLPTLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPM IILNTIMADSFDYMATALEKAVAGGVDFDSAVQTLLTEIITEHGAVVFNGDGYSENWQIEAAERGLPNLK TTLDAIPELIKPAAVEVFERYGVFSERELHSRYEVRLEQYALTIAVEAKLALELGTTVILPAAIRYQTEL AQNVATLKAAGMDADTTLLQAVSAPIAELTTALGALKAALSGHSGETALDEATHAQQALLPAMEAVRTAA DTLETVVADDLWPLPTYQEMLYIL*
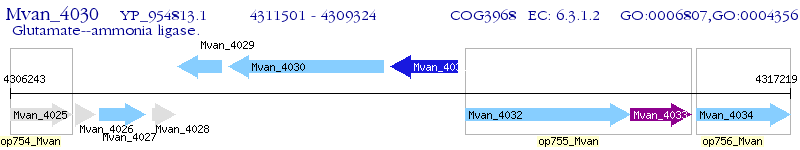
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4030 | - | - | 100% (725) | glutamine synthetase, catalytic region |
| M. vanbaalenii PYR-1 | Mvan_2920 | - | 2e-09 | 34.34% (99) | glutamine synthetase, catalytic region |
| M. vanbaalenii PYR-1 | Mvan_1850 | - | 5e-06 | 35.21% (71) | glutamate--ammonia ligase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2246c | glnA2 | 2e-05 | 30.61% (98) | glutamine synthetase |
| M. gilvum PYR-GCK | Mflv_2596 | - | 0.0 | 89.66% (725) | glutamine synthetase, catalytic region |
| M. tuberculosis H37Rv | Rv2222c | glnA2 | 2e-05 | 30.61% (98) | glutamine synthetase |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 2e-06 | 31.63% (98) | glutamine synthase class II |
| M. abscessus ATCC 19977 | MAB_1920 | - | 1e-05 | 30.63% (111) | glutamine synthetase |
| M. marinum M | MMAR_3549 | - | 0.0 | 88.14% (725) | hypothetical protein MMAR_3549 |
| M. avium 104 | MAV_2244 | glnA | 1e-06 | 25.61% (164) | glutamine synthetase, type I |
| M. smegmatis MC2 155 | MSMEG_4294 | glnA | 2e-06 | 27.94% (136) | glutamine synthetase, type I |
| M. thermoresistible (build 8) | TH_3396 | - | 3e-06 | 27.35% (117) | PUTATIVE glutamine synthetase, type I |
| M. ulcerans Agy99 | MUL_2782 | - | 0.0 | 87.03% (725) | hypothetical protein MUL_2782 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4294|M.smegmatis_MC2_155 ----------MDRQKEFVLR-------------------TLEERDIRFVR
TH_3396|M.thermoresistible__bu ----------MDRQKEFVLR-------------------SLEERDIRFVR
Mb2246c|M.bovis_AF2122/97 ----------MDRQKEFVLR-------------------TLEERDIRFVR
Rv2222c|M.tuberculosis_H37Rv ----------MDRQKEFVLR-------------------TLEERDIRFVR
MAV_2244|M.avium_104 ----------MDRQKEFVLR-------------------TLEERDIRFVR
MLBr_01631|M.leprae_Br4923 ----------MDRQKEFVLR-------------------TLEERDIRFVR
MAB_1920|M.abscessus_ATCC_1997 ----------MDRQKEFVLR-------------------TLEERDIRFVR
Mvan_4030|M.vanbaalenii_PYR-1 MSGNAVRLQAINNVEAYVPPAISFDPTEAPGQIFGSNVFTKAEMQLRLPK
Mflv_2596|M.gilvum_PYR-GCK MSGNAVRLQAIDNVEAYVPPAITFDPAEAPGEIFGSNVFTKAEMQSRLPK
MMAR_3549|M.marinum_M MSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPK
MUL_2782|M.ulcerans_Agy99 MSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPK
::. : :* : * : *: :
MSMEG_4294|M.smegmatis_MC2_155 LWFTDVLGYLKS---------VAIAPAELEGAFEEGIG------------
TH_3396|M.thermoresistible__bu LWFTDVLGFLKS---------VAIAPAELEGAFEEGIG------------
Mb2246c|M.bovis_AF2122/97 LWFTDVLGFLKS---------VAIAPAELEGAFEEGIG------------
Rv2222c|M.tuberculosis_H37Rv LWFTDVLGFLKS---------VAIAPAELEGAFEEGIG------------
MAV_2244|M.avium_104 LWFTDVLGFLKS---------VAIAPAELEGAFEEGIG------------
MLBr_01631|M.leprae_Br4923 LWFTDVLGYLKS---------VAIAPAELEGAFEEGIG------------
MAB_1920|M.abscessus_ATCC_1997 LWFTDVLGYLKS---------VSIAPAELEGAFEEGIG------------
Mvan_4030|M.vanbaalenii_PYR-1 SVYKSVVATIEKGAKLDPAIADAVASAMKDWALSKGATHYAHVFYPMTGL
Mflv_2596|M.gilvum_PYR-GCK SVYKSVVATIEKGVKLDPAVADSVASAMKDWALSKGATHYAHVFYPMTGF
MMAR_3549|M.marinum_M AVFKSVLATIDKGARLDPAVADAVAVAMKDWALEKGATHYAHVFYPMTGL
MUL_2782|M.ulcerans_Agy99 AVFKSVLATIDKGARLDPAVADAVAVAMKDWALEKGATHYAHVFYPMTGL
:..*:. :.. ::* * : *:.:*
MSMEG_4294|M.smegmatis_MC2_155 ------------FDGSSIEGFAR----VFESDTVARPDPSTFQVLPWKTS
TH_3396|M.thermoresistible__bu ------------FDGSAIEGFAR----VSEADMVARPDPSTFQVLPWTGD
Mb2246c|M.bovis_AF2122/97 ------------FDGSSIEGFAR----VSESDTVAHPDPSTFQVLPWATS
Rv2222c|M.tuberculosis_H37Rv ------------FDGSSIEGFAR----VSESDTVAHPDPSTFQVLPWATS
MAV_2244|M.avium_104 ------------FDGSSIEGFSR----VSESDTVANPDPSTFQVLPWASP
MLBr_01631|M.leprae_Br4923 ------------FDGSSIEGFAR----VSESDTVAHPDPSTFQILPWATT
MAB_1920|M.abscessus_ATCC_1997 ------------FDGSSIEGFAR----VSESDTVAHPDPSTFQVLPWTTS
Mvan_4030|M.vanbaalenii_PYR-1 TAEKHDSFLEPISDGETLAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
Mflv_2596|M.gilvum_PYR-GCK TAEKHDSFLEPVSDGQTLAEFAGKTLIQGEPDASSFPSGGLRTTFEARGY
MMAR_3549|M.marinum_M TAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
MUL_2782|M.ulcerans_Agy99 TAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
**.:: *: *.* : *. . :
MSMEG_4294|M.smegmatis_MC2_155 DGNHYSARMFCD--------------ITMPDGSPSWADSRHVLRRQLAKA
TH_3396|M.thermoresistible__bu SGKHYSARMFCD--------------ITMPDGSPSWADPRHVLRRQLAKA
Mb2246c|M.bovis_AF2122/97 SGHHHSARMFCD--------------ITMPDGSPSWADPRHVLRRQLTKA
Rv2222c|M.tuberculosis_H37Rv SGHHHSARMFCD--------------ITMPDGSPSWADPRHVLRRQLTKA
MAV_2244|M.avium_104 NGHHHSARMFCD--------------ITMPDGSPSWADPRHVLRRQLQKA
MLBr_01631|M.leprae_Br4923 AGHHHSARMFCD--------------ITMPDGSSSWADPRHVLRRQLTKA
MAB_1920|M.abscessus_ATCC_1997 AGKHHSARMFCD--------------IKLPDGSPSWSDPRHVLRRQLAKA
Mvan_4030|M.vanbaalenii_PYR-1 TGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDYKTPLLRSQQAMGVH
Mflv_2596|M.gilvum_PYR-GCK TGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDFKTPLLRSQQAMGAQ
MMAR_3549|M.marinum_M TGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIH
MUL_2782|M.ulcerans_Agy99 TGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIH
* . :: : :.:.. : .: . :: :
MSMEG_4294|M.smegmatis_MC2_155 SD-----------LGFTCYVHPEIEFFLLK------------PGPNDG--
TH_3396|M.thermoresistible__bu GD-----------QGFTCYVHPEIEFFLLK------------PGPHDG--
Mb2246c|M.bovis_AF2122/97 GE-----------LGFSCYVHPEIEFFLLK------------PGPEDG--
Rv2222c|M.tuberculosis_H37Rv GE-----------LGFSCYVHPEIEFFLLK------------PGPEDG--
MAV_2244|M.avium_104 ND-----------LGFSCYVHPEIEFFLLK------------PGPDDG--
MLBr_01631|M.leprae_Br4923 ND-----------LGFSCYVHPEIEFFLLK------------AGPEDGSM
MAB_1920|M.abscessus_ATCC_1997 SD-----------LGFSCYVHPEIEFFLLK------------NLPDDG--
Mvan_4030|M.vanbaalenii_PYR-1 AERILALFGHDNLQKVVSFCGPEQEYFLVDRHFFLARPDLVNAGRTLFGA
Mflv_2596|M.gilvum_PYR-GCK AERILKLFGHSDFDNIVSFCGPEQEYFLVDRHFFLARPDLINAGRTLFGT
MMAR_3549|M.marinum_M AERILKLFGHKELNKVVSFCGPEQEYFLVDRHFFVARPDLVNAGRTVFGA
MUL_2782|M.ulcerans_Agy99 AERILKLFGHEELNKVVSFCGPEQEYFLVDRHFFVARPELVNAGRTVFGA
: . .: ** *:**:.
MSMEG_4294|M.smegmatis_MC2_155 TPPEPADNGGYFDQAVHDAAPNFRRHAIEALEQMGISVEFSHHEGAPGQQ
TH_3396|M.thermoresistible__bu SPPVPADTSGYFDQSVHDTAPNFRRHAIEALEQMGISVEFSHHEGAPGQQ
Mb2246c|M.bovis_AF2122/97 SVPVPVDNAGYFDQAVHDSALNFRRHAIDALEFMGISVEFSHHEGAPGQQ
Rv2222c|M.tuberculosis_H37Rv SVPVPVDNAGYFDQAVHDSALNFRRHAIDALEFMGISVEFSHHEGAPGQQ
MAV_2244|M.avium_104 TPPVPVDTAGYFDQAIHDSASNFRRHAIEALEFMGISVEFSHHEGAPGQQ
MLBr_01631|M.leprae_Br4923 PIPVPVDNAGYFDQAVHDSALNFRRHAIEALEFMGISVEFSHHEGAPGQQ
MAB_1920|M.abscessus_ATCC_1997 TAPIPADDAGYFDQAVHEAAPNFRRHAIDALESMGISVEFSHHEGAPGQQ
Mvan_4030|M.vanbaalenii_PYR-1 KPPKGQEFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPAQF
Mflv_2596|M.gilvum_PYR-GCK RPPKGQEFDDHYFGAIPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
MMAR_3549|M.marinum_M KPPKGQEFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
MUL_2782|M.ulcerans_Agy99 KPPKGQGFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
* .:: :: : . .* .: * :**..: *:* **.*
MSMEG_4294|M.smegmatis_MC2_155 EIDLRYADALSMADNVMTFRYLVKEVALADGVRASFMPKPFAEHPGSAMH
TH_3396|M.thermoresistible__bu EIDLRYADALSMADNVMTFRYVVKQVAISDGVRASFMPKPFAEHPGSAMH
Mb2246c|M.bovis_AF2122/97 EIDLRFADALSMADNVMTFRYVIKEVALEEGARASFMPKPFGQHPGSAMH
Rv2222c|M.tuberculosis_H37Rv EIDLRFADALSMADNVMTFRYVIKEVALEEGARASFMPKPFGQHPGSAMH
MAV_2244|M.avium_104 EIDLRFADALSMADNVMTFRYVIKEVAIENGARASFMPKPFGQHPGSAMH
MLBr_01631|M.leprae_Br4923 EIDLRFADALSMADNVMTFRYVIKEVAIENGARASFMPKPFAQHPGSAMH
MAB_1920|M.abscessus_ATCC_1997 EIDLRYADALSMADNVMTFRNVVKEVAIIDGVHATFMPKPFSDHPGSAMH
Mvan_4030|M.vanbaalenii_PYR-1 EVAPMFERANIASDHQQLLMTVFKTVAKKHGMECLFHEKPFAGVNGSGKH
Mflv_2596|M.gilvum_PYR-GCK EIAPMFERANIAADHQQLLMTTFKTIAKKHGMECLFHEKPFAGVNGSGKH
MMAR_3549|M.marinum_M EVAPMFERANIASDHQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKH
MUL_2782|M.ulcerans_Agy99 EVAPMFERANIASDRQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKQ
*: : * :*. : .* :* .* .. * ***. **. :
MSMEG_4294|M.smegmatis_MC2_155 THMSLFEGDTNAFHSPDDPLQLSDVAKSFIAGILEHANEISAVT------
TH_3396|M.thermoresistible__bu THMSLFEGDTNAFHSPDDPLQLSDVAKSFIAGILEHANEISAVT------
Mb2246c|M.bovis_AF2122/97 THMSLFEGDVNAFHSADDPLQLSEVGKSFIAGILEHACEISAVT------
Rv2222c|M.tuberculosis_H37Rv THMSLFEGDVNAFHSADDPLQLSEVGKSFIAGILEHACEISAVT------
MAV_2244|M.avium_104 THMSLFEGDVNAFHSPDDPLQLSDVGKSFIAGILEHASEISAVT------
MLBr_01631|M.leprae_Br4923 THMSLFEGDVNAFHSDDDPLQLSDVGKSFIAGILEHASEISAVT------
MAB_1920|M.abscessus_ATCC_1997 THMSLFEGDTNAFHNPDDPLQLSDVGKSFIAGILEHANEISAVT------
Mvan_4030|M.vanbaalenii_PYR-1 VNFSVGNADLGSLLVPGDTPHENAQFLVFCAAIIRAVHKFGGLLRVSVAS
Mflv_2596|M.gilvum_PYR-GCK VNFSMGNSQFGSLLVPGDTPHENAQFLVFCAAVIRAVHKFAGLLRVSVAS
MMAR_3549|M.marinum_M VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAGLLRVSVAS
MUL_2782|M.ulcerans_Agy99 VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAELLRVSVAS
.::*: :.: .:: .*. : . * *.::. . : . :
MSMEG_4294|M.smegmatis_MC2_155 ----------------------NQWVNSYKRLVHGGEAPTAAS----WGA
TH_3396|M.thermoresistible__bu ----------------------NQWVNSYKRLVHGGEAPTAAS----WGA
Mb2246c|M.bovis_AF2122/97 ----------------------NQWVNSYKRLVQGGEAPTAAS----WGA
Rv2222c|M.tuberculosis_H37Rv ----------------------NQWVNSYKRLVQGGEAPTAAS----WGA
MAV_2244|M.avium_104 ----------------------NQWVNSYKRLVGGGEAPTAAS----WGA
MLBr_01631|M.leprae_Br4923 ----------------------NQWVNSYKRLVVGGEAPTTAS----WGA
MAB_1920|M.abscessus_ATCC_1997 ----------------------NQWVNSYKRLVQGGEAPTAAS----WGA
Mvan_4030|M.vanbaalenii_PYR-1 ATNDHRLGANEAPPAIISIFLGEQLADVFDQIAKGAATSSKGKGVMHIGV
Mflv_2596|M.gilvum_PYR-GCK ATNDHRLGANEAPPAIISIFLGDQLADVFEQIAKGAATSSKGKGSMIIGC
MMAR_3549|M.marinum_M ATNDHRLGANEAPPAIISIFLGEQLADVFEQIAKGAATSSKGKGTMIIGV
MUL_2782|M.ulcerans_Agy99 ATNDHRLGANEAPPAIISIFLGEQLADVFEQIAKGAATSSKGKGTMIIGV
:* .: :.::. *. :.: .. *
MSMEG_4294|M.smegmatis_MC2_155 ANRSALVRVP----MYTPHKVSSRRVEVRSPDS---ACNPYLTFAVLLAA
TH_3396|M.thermoresistible__bu ANRSALVRVP----MYTPHKASSRRVEVRSPDS---ACNPYLTFAVLLAA
Mb2246c|M.bovis_AF2122/97 ANRSALVRVP----MYTPHKTSSRRVEVRSPDS---ACNPYLTFAVLLAA
Rv2222c|M.tuberculosis_H37Rv ANRSALVRVP----MYTPHKTSSRRVEVRSPDS---ACNPYLTFAVLLAA
MAV_2244|M.avium_104 ANRSALVRVP----MYTPHKTSSRRVEVRSPDS---ACNPYLTFAVLLAA
MLBr_01631|M.leprae_Br4923 ANRSALVRVP----MYTPHKTSSRRVEVRSPDS---ACNPYLAFAVLLAA
MAB_1920|M.abscessus_ATCC_1997 ANRSALVRVP----MYTPNKSSSRRIEVRSPDS---ACNPYLTYAVLLAA
Mvan_4030|M.vanbaalenii_PYR-1 DTLPTLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMIILNTIMAD
Mflv_2596|M.gilvum_PYR-GCK DTLPILPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTINVPMIVLNTIMAD
MMAR_3549|M.marinum_M DTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMTILNTIMAD
MUL_2782|M.ulcerans_Agy99 DTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMTILNTIMAD
. . * * :* :..*.*.*:*.* * .::*
MSMEG_4294|M.smegmatis_MC2_155 GLR----GVEKGYVLGPQAEDNVWSLTQEERRAMGYRELPTSLGNALESM
TH_3396|M.thermoresistible__bu GLR----GVEKGYVLGPEAEDNVWTLTPEERRAMGFKELPSSLGQALAEM
Mb2246c|M.bovis_AF2122/97 GLR----GVEKGYVLGPQAEDNVWDLTPEERRAMGYRELPSSLDSALRAM
Rv2222c|M.tuberculosis_H37Rv GLR----GVEKGYVLGPQAEDNVWDLTPEERRAMGYRELPSSLDSALRAM
MAV_2244|M.avium_104 GLR----GVEKGYVLGPQAEDNVWDLTPEERIAMGYRELPTSLDSALRAM
MLBr_01631|M.leprae_Br4923 GLR----GVEKGYVLGPQAEDNVWDLTPEERLAMGYRELPTSLDSALGAM
MAB_1920|M.abscessus_ATCC_1997 GLR----GVEQGYTLGPEAEDDVWGLTRAERQAMGYKELPSTLENALIAM
Mvan_4030|M.vanbaalenii_PYR-1 SFDYMATALEKAVAGGVDFDSAVQTLLTEIITEHGAVVFNGDGYSENWQI
Mflv_2596|M.gilvum_PYR-GCK SLDYMATVLETAVGDGADFDTAVQKLLTEIITEHGAVVFNGDGYSENWHI
MMAR_3549|M.marinum_M SFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQI
MUL_2782|M.ulcerans_Agy99 SFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQV
.: :* . * : : * * * : . :
MSMEG_4294|M.smegmatis_MC2_155 ENSE-----------LVAEALGEHVFDYFLR----NKRSEWENYRSHVTP
TH_3396|M.thermoresistible__bu ERSE-----------LVAEALGEHVFDFFLR----NKRAEWENYRSHVTP
Mb2246c|M.bovis_AF2122/97 EASE-----------LVAEALGEHVFDFFLR----NKRTEWANYRSHVTP
Rv2222c|M.tuberculosis_H37Rv EASE-----------LVAEALGEHVFDFFLR----NKRTEWANYRSHVTP
MAV_2244|M.avium_104 ESSE-----------LVAETLGEHVFDFFLR----NKRTEWATYRSHVTP
MLBr_01631|M.leprae_Br4923 EVSE-----------FVAEALGEHVFDFFLR----NKRAEWANYRSHVSL
MAB_1920|M.abscessus_ATCC_1997 EGSE-----------LVAEALGEHVFDFFLR----NKRAEWARYRSNVTP
Mvan_4030|M.vanbaalenii_PYR-1 EAAERGLPNLKTTLDAIPELIKPAAVEVFERYGVFSERELHSRYEVRLEQ
Mflv_2596|M.gilvum_PYR-GCK EAAERGLPNLKTTLDAITELIKPEAVEVFEKYGVFSERELHSRYEVRLEQ
MMAR_3549|M.marinum_M EAAERGLPNLKTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQ
MUL_2782|M.ulcerans_Agy99 EAAERGLPNLKTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQ
* :* :.* : .: * : .:* *. .:
MSMEG_4294|M.smegmatis_MC2_155 YELKNYLSL-----------------------------------------
TH_3396|M.thermoresistible__bu YELKNYLSL-----------------------------------------
Mb2246c|M.bovis_AF2122/97 YELRTYLSL-----------------------------------------
Rv2222c|M.tuberculosis_H37Rv YELRTYLSL-----------------------------------------
MAV_2244|M.avium_104 YELRTYLSL-----------------------------------------
MLBr_01631|M.leprae_Br4923 YELKTYLSL-----------------------------------------
MAB_1920|M.abscessus_ATCC_1997 FELRAYLGL-----------------------------------------
Mvan_4030|M.vanbaalenii_PYR-1 YALTIAVEAKLALELGTTVILPAAIRYQTELAQNVATLKAAGMDADTTLL
Mflv_2596|M.gilvum_PYR-GCK YALTIAVEAKLSLELGTTMILPAAVRYQTELAQNVAALKAAGVAADTTLL
MMAR_3549|M.marinum_M YALTIAVEARLTLEMGTTVILPAAMRYQTELAQNVAALKAAGVEPSMVAL
MUL_2782|M.ulcerans_Agy99 YALTIAVEARLTLEMGTTVILPAAMRYQTELAQNVAALKAAGVEPSMVAL
: * :
MSMEG_4294|M.smegmatis_MC2_155 --------------------------------------------------
TH_3396|M.thermoresistible__bu --------------------------------------------------
Mb2246c|M.bovis_AF2122/97 --------------------------------------------------
Rv2222c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_2244|M.avium_104 --------------------------------------------------
MLBr_01631|M.leprae_Br4923 --------------------------------------------------
MAB_1920|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_4030|M.vanbaalenii_PYR-1 QAVSAPIAELTTALGALKAALSGHSGETALDEATHAQQALLPAMEAVRTA
Mflv_2596|M.gilvum_PYR-GCK QEVSAPIAELTAALATLKSALSGHSAESALDEATHAQKALLPAMEAVRAA
MMAR_3549|M.marinum_M EAISAPLSDLSAALASLKMALSEHSADSALSEAKHAQEALLPAMDAVRCA
MUL_2782|M.ulcerans_Agy99 EAISAPLSDLSAALASLKMALSEHSADSALSDAKHAQEALLPAMDAVRCA
MSMEG_4294|M.smegmatis_MC2_155 -------------------------
TH_3396|M.thermoresistible__bu -------------------------
Mb2246c|M.bovis_AF2122/97 -------------------------
Rv2222c|M.tuberculosis_H37Rv -------------------------
MAV_2244|M.avium_104 -------------------------
MLBr_01631|M.leprae_Br4923 -------------------------
MAB_1920|M.abscessus_ATCC_1997 -------------------------
Mvan_4030|M.vanbaalenii_PYR-1 ADTLETVVADDLWPLPTYQEMLYIL
Mflv_2596|M.gilvum_PYR-GCK ADELEAVVADDLWPLPTYQEMLYIL
MMAR_3549|M.marinum_M ADTLESVVADDLWPLPTYQEMLYIL
MUL_2782|M.ulcerans_Agy99 ADTLESVVADDLWPLPTYQKMLYIL