For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSGNAVRLQAIDNVEAYVPPAITFDPAEAPGEIFGSNVFTKAEMQSRLPKSVYKSVVATIEKGVKLDPAV ADSVASAMKDWALSKGATHYAHVFYPMTGFTAEKHDSFLEPVSDGQTLAEFAGKTLIQGEPDASSFPSGG LRTTFEARGYTGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDFKTPLLRSQQAMGAQAERILKLFGH SDFDNIVSFCGPEQEYFLVDRHFFLARPDLINAGRTLFGTRPPKGQEFDDHYFGAIPERVLGFMMDTERE LFKLGIPAKTRHNEVAPGQFEIAPMFERANIAADHQQLLMTTFKTIAKKHGMECLFHEKPFAGVNGSGKH VNFSMGNSQFGSLLVPGDTPHENAQFLVFCAAVIRAVHKFAGLLRVSVASATNDHRLGANEAPPAIISIF LGDQLADVFEQIAKGAATSSKGKGSMIIGCDTLPILPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTINVP MIVLNTIMADSLDYMATVLETAVGDGADFDTAVQKLLTEIITEHGAVVFNGDGYSENWHIEAAERGLPNL KTTLDAITELIKPEAVEVFEKYGVFSERELHSRYEVRLEQYALTIAVEAKLSLELGTTMILPAAVRYQTE LAQNVAALKAAGVAADTTLLQEVSAPIAELTAALATLKSALSGHSAESALDEATHAQKALLPAMEAVRAA ADELEAVVADDLWPLPTYQEMLYIL
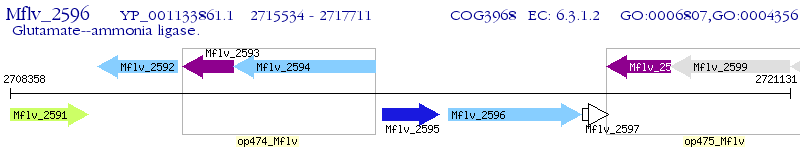
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2596 | - | - | 100% (725) | glutamine synthetase, catalytic region |
| M. gilvum PYR-GCK | Mflv_3319 | - | 4e-07 | 30.77% (130) | glutamine synthetase, catalytic region |
| M. gilvum PYR-GCK | Mflv_2909 | - | 3e-06 | 24.65% (142) | glutamine synthetase, type I |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1910 | glnA3 | 1e-05 | 29.23% (130) | glutamine synthetase |
| M. tuberculosis H37Rv | Rv1878 | glnA3 | 1e-05 | 29.23% (130) | glutamine synthetase |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 2e-06 | 32.97% (91) | glutamine synthase class II |
| M. abscessus ATCC 19977 | MAB_1920 | - | 3e-05 | 29.73% (111) | glutamine synthetase |
| M. marinum M | MMAR_3549 | - | 0.0 | 86.34% (725) | hypothetical protein MMAR_3549 |
| M. avium 104 | MAV_2244 | glnA | 1e-06 | 25.61% (164) | glutamine synthetase, type I |
| M. smegmatis MC2 155 | MSMEG_4294 | glnA | 3e-06 | 27.35% (117) | glutamine synthetase, type I |
| M. thermoresistible (build 8) | TH_3396 | - | 4e-06 | 26.50% (117) | PUTATIVE glutamine synthetase, type I |
| M. ulcerans Agy99 | MUL_2782 | - | 0.0 | 85.24% (725) | hypothetical protein MUL_2782 |
| M. vanbaalenii PYR-1 | Mvan_4030 | - | 0.0 | 89.66% (725) | glutamine synthetase, catalytic region |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4294|M.smegmatis_MC2_155 --------------------------------------------------
TH_3396|M.thermoresistible__bu --------------------------------------------------
MLBr_01631|M.leprae_Br4923 --------------------------------------------------
MAV_2244|M.avium_104 --------------------------------------------------
MAB_1920|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1910|M.bovis_AF2122/97 --------------------------------------------------
Rv1878|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_2596|M.gilvum_PYR-GCK MSGNAVRLQAIDNVEAYVPPAITFDPAEAPGEIFGSNVFTKAEMQSRLPK
Mvan_4030|M.vanbaalenii_PYR-1 MSGNAVRLQAINNVEAYVPPAISFDPTEAPGQIFGSNVFTKAEMQLRLPK
MMAR_3549|M.marinum_M MSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPK
MUL_2782|M.ulcerans_Agy99 MSGNAVRLQAINNVEAYVPPAVSFVAGEAPGEIFGSNVFTKAEMQARLPK
MSMEG_4294|M.smegmatis_MC2_155 ------------------------MDRQKEFVLRT---------------
TH_3396|M.thermoresistible__bu ------------------------MDRQKEFVLRS---------------
MLBr_01631|M.leprae_Br4923 ------------------------MDRQKEFVLRT---------------
MAV_2244|M.avium_104 ------------------------MDRQKEFVLRT---------------
MAB_1920|M.abscessus_ATCC_1997 ------------------------MDRQKEFVLRT---------------
Mb1910|M.bovis_AF2122/97 ----------------------MTATPLAAAAIAQ---------------
Rv1878|M.tuberculosis_H37Rv ----------------------MTATPLAAAAIAQ---------------
Mflv_2596|M.gilvum_PYR-GCK SVYKSVVATIEKGVKLDPAVADSVASAMKDWALSKGATHYAHVFYPMTGF
Mvan_4030|M.vanbaalenii_PYR-1 SVYKSVVATIEKGAKLDPAIADAVASAMKDWALSKGATHYAHVFYPMTGL
MMAR_3549|M.marinum_M AVFKSVLATIDKGARLDPAVADAVAVAMKDWALEKGATHYAHVFYPMTGL
MUL_2782|M.ulcerans_Agy99 AVFKSVLATIDKGARLDPAVADAVAVAMKDWALEKGATHYAHVFYPMTGL
.:
MSMEG_4294|M.smegmatis_MC2_155 -LEERDIRFVRLWFTDVLGYLKSVAIAPAELEGAFEEGIG----FDGSSI
TH_3396|M.thermoresistible__bu -LEERDIRFVRLWFTDVLGFLKSVAIAPAELEGAFEEGIG----FDGSAI
MLBr_01631|M.leprae_Br4923 -LEERDIRFVRLWFTDVLGYLKSVAIAPAELEGAFEEGIG----FDGSSI
MAV_2244|M.avium_104 -LEERDIRFVRLWFTDVLGFLKSVAIAPAELEGAFEEGIG----FDGSSI
MAB_1920|M.abscessus_ATCC_1997 -LEERDIRFVRLWFTDVLGYLKSVSIAPAELEGAFEEGIG----FDGSSI
Mb1910|M.bovis_AF2122/97 -LEAEGVDTVIGTVVNPAGLTQAKTVPIRRTNTFANPGLGASPVWHTFCI
Rv1878|M.tuberculosis_H37Rv -LEAEGVDTVIGTVVNPAGLTQAKTVPIRRTNTFANPGLGASPVWHTFCI
Mflv_2596|M.gilvum_PYR-GCK TAEKHDSFLEPVSDGQTLAEFAGKTLIQGEPDASSFPSGGLRTTFEARGY
Mvan_4030|M.vanbaalenii_PYR-1 TAEKHDSFLEPISDGETLAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
MMAR_3549|M.marinum_M TAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
MUL_2782|M.ulcerans_Agy99 TAEKHDSFLEPTADGQTMAEFAGKTLIQGEPDASSFPSGGLRSTFEARGY
* .. : . . :: . : . * :.
MSMEG_4294|M.smegmatis_MC2_155 EGFARVFESDTVARPDPSTFQVLPWKTSDGNHYSARMFCDITMPDGSPSW
TH_3396|M.thermoresistible__bu EGFARVSEADMVARPDPSTFQVLPWTGDSGKHYSARMFCDITMPDGSPSW
MLBr_01631|M.leprae_Br4923 EGFARVSESDTVAHPDPSTFQILPWATTAGHHHSARMFCDITMPDGSSSW
MAV_2244|M.avium_104 EGFSRVSESDTVANPDPSTFQVLPWASPNGHHHSARMFCDITMPDGSPSW
MAB_1920|M.abscessus_ATCC_1997 EGFARVSESDTVAHPDPSTFQVLPWTTSAGKHHSARMFCDIKLPDGSPSW
Mb1910|M.bovis_AF2122/97 DQCSIAFTADISVVGDQRLRIDLSALRIIG-DGLAWAPAGFFEQDGTPVP
Rv1878|M.tuberculosis_H37Rv DQCSIAFTADISVVGDQRLRIDLSALRIIG-DGLAWAPAGFFEQDGTPVP
Mflv_2596|M.gilvum_PYR-GCK TGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDFKTPLLRSQQAMGAQ
Mvan_4030|M.vanbaalenii_PYR-1 TGWDVTSPAYILENPNGNTLCIPTVFVSMTGEALDYKTPLLRSQQAMGVH
MMAR_3549|M.marinum_M TGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIH
MUL_2782|M.ulcerans_Agy99 TGWDVTSPAYVLENPNGNTLCIPTIFVSMTGDALDYKTPLLRSQQAMGIH
. : : . . : :.
MSMEG_4294|M.smegmatis_MC2_155 ADSRHVLR-----RQLAKASDLGFTCYVHPEIEFFLLKPGPNDGTP----
TH_3396|M.thermoresistible__bu ADPRHVLR-----RQLAKAGDQGFTCYVHPEIEFFLLKPGPHDGSP----
MLBr_01631|M.leprae_Br4923 ADPRHVLR-----RQLTKANDLGFSCYVHPEIEFFLLKAGPEDGSMPI--
MAV_2244|M.avium_104 ADPRHVLR-----RQLQKANDLGFSCYVHPEIEFFLLKPGPDDGTP----
MAB_1920|M.abscessus_ATCC_1997 SDPRHVLR-----RQLAKASDLGFSCYVHPEIEFFLLKNLPDDGTA----
Mb1910|M.bovis_AF2122/97 ACSRGTLS-----RIEAALADAGIDAVIGHEVEFLLVD---ADGQR----
Rv1878|M.tuberculosis_H37Rv ACSRGTLS-----RIEAALADAGIDAVIGHEVEFLLVD---ADGQR----
Mflv_2596|M.gilvum_PYR-GCK AERILKLFGHSDFDNIVSFCGPEQEYFLVDRHFFLARPDLINAGRTLFGT
Mvan_4030|M.vanbaalenii_PYR-1 AERILALFGHDNLQKVVSFCGPEQEYFLVDRHFFLARPDLVNAGRTLFGA
MMAR_3549|M.marinum_M AERILKLFGHKELNKVVSFCGPEQEYFLVDRHFFVARPDLVNAGRTVFGA
MUL_2782|M.ulcerans_Agy99 AERILKLFGHEELNKVVSFCGPEQEYFLVDRHFFVARPELVNAGRTVFGA
: * . : . *. *
MSMEG_4294|M.smegmatis_MC2_155 --PEPADNGGYFDQAVHDAAPNFRRHAIEALEQMGISVEFSHHEGAPGQQ
TH_3396|M.thermoresistible__bu --PVPADTSGYFDQSVHDTAPNFRRHAIEALEQMGISVEFSHHEGAPGQQ
MLBr_01631|M.leprae_Br4923 --PVPVDNAGYFDQAVHDSALNFRRHAIEALEFMGISVEFSHHEGAPGQQ
MAV_2244|M.avium_104 --PVPVDTAGYFDQAIHDSASNFRRHAIEALEFMGISVEFSHHEGAPGQQ
MAB_1920|M.abscessus_ATCC_1997 --PIPADDAGYFDQAVHEAAPNFRRHAIDALESMGISVEFSHHEGAPGQQ
Mb1910|M.bovis_AF2122/97 ---LPSTLWAQYGVAGVLEHEAFVRDVNAAATAAGIAIEQFHPEYGANQF
Rv1878|M.tuberculosis_H37Rv ---LPSTLWAQYGVAGVLEHEAFVRDVNAAATAAGIAIEQFHPEYGANQF
Mflv_2596|M.gilvum_PYR-GCK RPPKGQEFDDHYFGAIPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
Mvan_4030|M.vanbaalenii_PYR-1 KPPKGQEFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPAQF
MMAR_3549|M.marinum_M KPPKGQEFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
MUL_2782|M.ulcerans_Agy99 KPPKGQGFDDHYFGAVPERVLGFMMDTERELFKLGIPAKTRHNEVAPGQF
: : * .. **. : * * .. *
MSMEG_4294|M.smegmatis_MC2_155 EIDLRYADALSMADNVMTFRYLVKEVALADGVRASFMPKPFAEHPGSAMH
TH_3396|M.thermoresistible__bu EIDLRYADALSMADNVMTFRYVVKQVAISDGVRASFMPKPFAEHPGSAMH
MLBr_01631|M.leprae_Br4923 EIDLRFADALSMADNVMTFRYVIKEVAIENGARASFMPKPFAQHPGSAMH
MAV_2244|M.avium_104 EIDLRFADALSMADNVMTFRYVIKEVAIENGARASFMPKPFGQHPGSAMH
MAB_1920|M.abscessus_ATCC_1997 EIDLRYADALSMADNVMTFRNVVKEVAIIDGVHATFMPKPFSDHPGSAMH
Mb1910|M.bovis_AF2122/97 EISLAPQPPVAAADQLVLTRLIIGRTARRHGLRVSLSPAPFAGSIGSGAH
Rv1878|M.tuberculosis_H37Rv EISLAPQPPVAAADQLVLTRLIIGRTARRHGLRVSLSPAPFAGSIGSGAH
Mflv_2596|M.gilvum_PYR-GCK EIAPMFERANIAADHQQLLMTTFKTIAKKHGMECLFHEKPFAGVNGSGKH
Mvan_4030|M.vanbaalenii_PYR-1 EVAPMFERANIASDHQQLLMTVFKTVAKKHGMECLFHEKPFAGVNGSGKH
MMAR_3549|M.marinum_M EVAPMFERANIASDHQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKH
MUL_2782|M.ulcerans_Agy99 EVAPMFERANIASDRQQLLMTVFKTLAKKHGMECLFHEKPFAGVNGSGKQ
*: . :*. . * .* . : **. **. :
MSMEG_4294|M.smegmatis_MC2_155 THMSLFEGDTNAFHSPDDPLQLSDVAKSFIAGILEHANEISAVTNQWVNS
TH_3396|M.thermoresistible__bu THMSLFEGDTNAFHSPDDPLQLSDVAKSFIAGILEHANEISAVTNQWVNS
MLBr_01631|M.leprae_Br4923 THMSLFEGDVNAFHSDDDPLQLSDVGKSFIAGILEHASEISAVTNQWVNS
MAV_2244|M.avium_104 THMSLFEGDVNAFHSPDDPLQLSDVGKSFIAGILEHASEISAVTNQWVNS
MAB_1920|M.abscessus_ATCC_1997 THMSLFEGDTNAFHNPDDPLQLSDVGKSFIAGILEHANEISAVTNQWVNS
Mb1910|M.bovis_AF2122/97 QHFSLTMSEGMLFSGGTGAAGMTSAGEATVAGVLRGLPDAQGILCGSIVS
Rv1878|M.tuberculosis_H37Rv QHFSLTMSEGMLFSGGTGAAGMTSAGEAAVAGVLRGLPDAQGILCGSIVS
Mflv_2596|M.gilvum_PYR-GCK VNFSMGNSQFGSLLVPGDTPHENAQFLVFCAAVIRAVHKFAGLLRVSVAS
Mvan_4030|M.vanbaalenii_PYR-1 VNFSVGNADLGSLLVPGDTPHENAQFLVFCAAIIRAVHKFGGLLRVSVAS
MMAR_3549|M.marinum_M VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAGLLRVSVAS
MUL_2782|M.ulcerans_Agy99 VNFSVGNAELGSLLVPGDTPHENAQFLVFCAAVIRAVHKYAELLRVSVAS
::*: .: : .. . *.::. . : : *
MSMEG_4294|M.smegmatis_MC2_155 YKRLVHGGE-----APTAASWGAANRSALVRVPMYTPHKV----------
TH_3396|M.thermoresistible__bu YKRLVHGGE-----APTAASWGAANRSALVRVPMYTPHKA----------
MLBr_01631|M.leprae_Br4923 YKRLVVGGE-----APTTASWGAANRSALVRVPMYTPHKT----------
MAV_2244|M.avium_104 YKRLVGGGE-----APTAASWGAANRSALVRVPMYTPHKT----------
MAB_1920|M.abscessus_ATCC_1997 YKRLVQGGE-----APTAASWGAANRSALVRVPMYTPNKS----------
Mb1910|M.bovis_AF2122/97 GLRMRPGNW-----AGIYACWGTENREAAVRFVKGGAGSA----------
Rv1878|M.tuberculosis_H37Rv GLRMRPGNW-----AGIYACWGTENREAAVRFVKGGAGSA----------
Mflv_2596|M.gilvum_PYR-GCK ATNDHRLGANEAPPAIISIFLGDQLADVFEQIAKGAATSSKGKGSMIIGC
Mvan_4030|M.vanbaalenii_PYR-1 ATNDHRLGANEAPPAIISIFLGEQLADVFDQIAKGAATSSKGKGVMHIGV
MMAR_3549|M.marinum_M ATNDHRLGANEAPPAIISIFLGEQLADVFEQIAKGAATSSKGKGTMIIGV
MUL_2782|M.ulcerans_Agy99 ATNDHRLGANEAPPAIISIFLGEQLADVFEQIAKGAATSSKGKGTMIIGV
. . * * .. :. . .
MSMEG_4294|M.smegmatis_MC2_155 ---------------------SSRRVEVRSPDS--------------ACN
TH_3396|M.thermoresistible__bu ---------------------SSRRVEVRSPDS--------------ACN
MLBr_01631|M.leprae_Br4923 ---------------------SSRRVEVRSPDS--------------ACN
MAV_2244|M.avium_104 ---------------------SSRRVEVRSPDS--------------ACN
MAB_1920|M.abscessus_ATCC_1997 ---------------------SSRRIEVRSPDS--------------ACN
Mb1910|M.bovis_AF2122/97 ---------------------YGGNVEVKVVDP--------------SAN
Rv1878|M.tuberculosis_H37Rv ---------------------YGGNVEVKVVDP--------------SAN
Mflv_2596|M.gilvum_PYR-GCK DTLPILPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTINVPMIVLNTIMAD
Mvan_4030|M.vanbaalenii_PYR-1 DTLPTLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMIILNTIMAD
MMAR_3549|M.marinum_M DTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMTILNTIMAD
MUL_2782|M.ulcerans_Agy99 DTLPPLPTDPGDRNRTSPFAFTGNRFEFRAPGSGQTVAVPMTILNTIMAD
. ..*.: .. .:
MSMEG_4294|M.smegmatis_MC2_155 PYLTFAVLLAAGLRG-----------------VEKGYVLGPQAEDNVWSL
TH_3396|M.thermoresistible__bu PYLTFAVLLAAGLRG-----------------VEKGYVLGPEAEDNVWTL
MLBr_01631|M.leprae_Br4923 PYLAFAVLLAAGLRG-----------------VEKGYVLGPQAEDNVWDL
MAV_2244|M.avium_104 PYLTFAVLLAAGLRG-----------------VEKGYVLGPQAEDNVWDL
MAB_1920|M.abscessus_ATCC_1997 PYLTYAVLLAAGLRG-----------------VEQGYTLGPEAEDDVWGL
Mb1910|M.bovis_AF2122/97 PYLASAAILGLALDG-----------------MKTKAVLPSETTVDPTQL
Rv1878|M.tuberculosis_H37Rv PYLASAAILGLALDG-----------------MKTKAVLPSETTVDPTQL
Mflv_2596|M.gilvum_PYR-GCK SLDYMATVLETAVGDGADFDTAVQKLLTEIITEHGAVVFNGDGYSENWHI
Mvan_4030|M.vanbaalenii_PYR-1 SFDYMATALEKAVAGGVDFDSAVQTLLTEIITEHGAVVFNGDGYSENWQI
MMAR_3549|M.marinum_M SFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQI
MUL_2782|M.ulcerans_Agy99 SFDYMATVLEKAVEDGEDFDAAVQKLLTEIITEHGAVVFNGDGYSENWQV
. *. * .: . . .: : : :
MSMEG_4294|M.smegmatis_MC2_155 TQEER-------------------------------RAMGYRELPTSLGN
TH_3396|M.thermoresistible__bu TPEER-------------------------------RAMGFKELPSSLGQ
MLBr_01631|M.leprae_Br4923 TPEER-------------------------------LAMGYRELPTSLDS
MAV_2244|M.avium_104 TPEER-------------------------------IAMGYRELPTSLDS
MAB_1920|M.abscessus_ATCC_1997 TRAER-------------------------------QAMGYKELPSTLEN
Mb1910|M.bovis_AF2122/97 SDVDR-------------------------------DRAGILRLAADQAD
Rv1878|M.tuberculosis_H37Rv SDVDR-------------------------------DRAGILRLAADQAD
Mflv_2596|M.gilvum_PYR-GCK EAAERGLPNLKTTLDAITELIKPEAVEVFEKYGVFSERELHSRYEVRLEQ
Mvan_4030|M.vanbaalenii_PYR-1 EAAERGLPNLKTTLDAIPELIKPAAVEVFERYGVFSERELHSRYEVRLEQ
MMAR_3549|M.marinum_M EAAERGLPNLKTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQ
MUL_2782|M.ulcerans_Agy99 EAAERGLPNLKTTLDAIPELIKPESIDLFHKYDVFNERELHSRYEVRLEQ
:* . .
MSMEG_4294|M.smegmatis_MC2_155 ALESMENSELVAEALGEHVFDYFLRNKRSEWENYRSHVTPYELKNYLSL-
TH_3396|M.thermoresistible__bu ALAEMERSELVAEALGEHVFDFFLRNKRAEWENYRSHVTPYELKNYLSL-
MLBr_01631|M.leprae_Br4923 ALGAMEVSEFVAEALGEHVFDFFLRNKRAEWANYRSHVSLYELKTYLSL-
MAV_2244|M.avium_104 ALRAMESSELVAETLGEHVFDFFLRNKRTEWATYRSHVTPYELRTYLSL-
MAB_1920|M.abscessus_ATCC_1997 ALIAMEGSELVAEALGEHVFDFFLRNKRAEWARYRSNVTPFELRAYLGL-
Mb1910|M.bovis_AF2122/97 AIAVLDSSKLLRCILGDPVVDAVVAVRQLEHERYG-DLDPAQLADKFRMA
Rv1878|M.tuberculosis_H37Rv AIAVLDSSKLLRCILGDPVVDAVVAVRQLEHERYG-DLDPAQLADKFRMA
Mflv_2596|M.gilvum_PYR-GCK YALTIAVEAKLSLELGTTMILPAAVRYQTELAQNVAALKAAGVAADTTLL
Mvan_4030|M.vanbaalenii_PYR-1 YALTIAVEAKLALELGTTVILPAAIRYQTELAQNVATLKAAGMDADTTLL
MMAR_3549|M.marinum_M YALTIAVEARLTLEMGTTVILPAAMRYQTELAQNVAALKAAGVEPSMVAL
MUL_2782|M.ulcerans_Agy99 YALTIAVEARLTLEMGTTVILPAAMRYQTELAQNVAALKAAGVEPSMVAL
: . : :* :. : * : :
MSMEG_4294|M.smegmatis_MC2_155 --------------------------------------------------
TH_3396|M.thermoresistible__bu --------------------------------------------------
MLBr_01631|M.leprae_Br4923 --------------------------------------------------
MAV_2244|M.avium_104 --------------------------------------------------
MAB_1920|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1910|M.bovis_AF2122/97 WSV-----------------------------------------------
Rv1878|M.tuberculosis_H37Rv WSV-----------------------------------------------
Mflv_2596|M.gilvum_PYR-GCK QEVSAPIAELTAALATLKSALSGHSAESALDEATHAQKALLPAMEAVRAA
Mvan_4030|M.vanbaalenii_PYR-1 QAVSAPIAELTTALGALKAALSGHSGETALDEATHAQQALLPAMEAVRTA
MMAR_3549|M.marinum_M EAISAPLSDLSAALASLKMALSEHSADSALSEAKHAQEALLPAMDAVRCA
MUL_2782|M.ulcerans_Agy99 EAISAPLSDLSAALASLKMALSEHSADSALSDAKHAQEALLPAMDAVRCA
MSMEG_4294|M.smegmatis_MC2_155 -------------------------
TH_3396|M.thermoresistible__bu -------------------------
MLBr_01631|M.leprae_Br4923 -------------------------
MAV_2244|M.avium_104 -------------------------
MAB_1920|M.abscessus_ATCC_1997 -------------------------
Mb1910|M.bovis_AF2122/97 -------------------------
Rv1878|M.tuberculosis_H37Rv -------------------------
Mflv_2596|M.gilvum_PYR-GCK ADELEAVVADDLWPLPTYQEMLYIL
Mvan_4030|M.vanbaalenii_PYR-1 ADTLETVVADDLWPLPTYQEMLYIL
MMAR_3549|M.marinum_M ADTLESVVADDLWPLPTYQEMLYIL
MUL_2782|M.ulcerans_Agy99 ADTLESVVADDLWPLPTYQKMLYIL