For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DRQKEFVLRTLEERDIRFVRLWFTDVLGYLKSVAIAPAELEGAFEEGIGFDGSSIEGFARVFESDTVARP DPSTFQVLPWKTSDGNHYSARMFCDITMPDGSPSWADSRHVLRRQLAKASDLGFTCYVHPEIEFFLLKPG PNDGTPPEPADNGGYFDQAVHDAAPNFRRHAIEALEQMGISVEFSHHEGAPGQQEIDLRYADALSMADNV MTFRYLVKEVALADGVRASFMPKPFAEHPGSAMHTHMSLFEGDTNAFHSPDDPLQLSDVAKSFIAGILEH ANEISAVTNQWVNSYKRLVHGGEAPTAASWGAANRSALVRVPMYTPHKVSSRRVEVRSPDSACNPYLTFA VLLAAGLRGVEKGYVLGPQAEDNVWSLTQEERRAMGYRELPTSLGNALESMENSELVAEALGEHVFDYFL RNKRSEWENYRSHVTPYELKNYLSL*
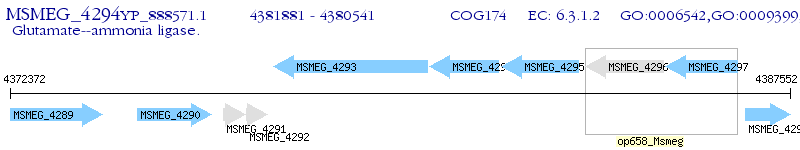
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4294 | glnA | - | 100% (446) | glutamine synthetase, type I |
| M. smegmatis MC2 155 | MSMEG_4290 | glnA | 8e-57 | 30.90% (479) | glutamine synthetase, type I |
| M. smegmatis MC2 155 | MSMEG_6260 | glnT | 6e-52 | 33.11% (450) | glutamine synthetase, type III |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2246c | glnA2 | 0.0 | 88.79% (446) | glutamine synthetase |
| M. gilvum PYR-GCK | Mflv_2909 | - | 0.0 | 93.05% (446) | glutamine synthetase, type I |
| M. tuberculosis H37Rv | Rv2222c | glnA2 | 0.0 | 88.79% (446) | glutamine synthetase |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 0.0 | 87.50% (448) | glutamine synthase class II |
| M. abscessus ATCC 19977 | MAB_1920 | - | 0.0 | 86.32% (446) | glutamine synthetase |
| M. marinum M | MMAR_3294 | glnA2 | 0.0 | 87.00% (446) | glutamine synthetase GlnA2 |
| M. avium 104 | MAV_2244 | glnA | 0.0 | 88.34% (446) | glutamine synthetase, type I |
| M. thermoresistible (build 8) | TH_3396 | - | 0.0 | 91.48% (446) | PUTATIVE glutamine synthetase, type I |
| M. ulcerans Agy99 | MUL_1335 | glnA2 | 0.0 | 86.77% (446) | glutamine synthetase GlnA2 |
| M. vanbaalenii PYR-1 | Mvan_3598 | - | 0.0 | 94.84% (446) | glutamine synthetase, type I |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2909|M.gilvum_PYR-GCK MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKSVAIAPAELEGAFEEGIG
Mvan_3598|M.vanbaalenii_PYR-1 MDRQMEFVLRTLEERDIRFVRLWFTDVLGYLKSVAIAPAELEGAFEEGIG
MSMEG_4294|M.smegmatis_MC2_155 MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKSVAIAPAELEGAFEEGIG
TH_3396|M.thermoresistible__bu MDRQKEFVLRSLEERDIRFVRLWFTDVLGFLKSVAIAPAELEGAFEEGIG
Mb2246c|M.bovis_AF2122/97 MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAIAPAELEGAFEEGIG
Rv2222c|M.tuberculosis_H37Rv MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAIAPAELEGAFEEGIG
MMAR_3294|M.marinum_M MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAIAPAELEGAFEEGIG
MUL_1335|M.ulcerans_Agy99 MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAIAPAELEGAFEEGIG
MAV_2244|M.avium_104 MDRQKEFVLRTLEERDIRFVRLWFTDVLGFLKSVAIAPAELEGAFEEGIG
MLBr_01631|M.leprae_Br4923 MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKSVAIAPAELEGAFEEGIG
MAB_1920|M.abscessus_ATCC_1997 MDRQKEFVLRTLEERDIRFVRLWFTDVLGYLKSVSIAPAELEGAFEEGIG
**** *****:******************:****:***************
Mflv_2909|M.gilvum_PYR-GCK FDGSSIEGFARVSEADMVARPDPSTFQVLPWADSSGKHHSARMFCDITMP
Mvan_3598|M.vanbaalenii_PYR-1 FDGSSIEGFARVSEADMVARPDPSTFQVLPWTTSAGNHYSARMFCDITMP
MSMEG_4294|M.smegmatis_MC2_155 FDGSSIEGFARVFESDTVARPDPSTFQVLPWKTSDGNHYSARMFCDITMP
TH_3396|M.thermoresistible__bu FDGSAIEGFARVSEADMVARPDPSTFQVLPWTGDSGKHYSARMFCDITMP
Mb2246c|M.bovis_AF2122/97 FDGSSIEGFARVSESDTVAHPDPSTFQVLPWATSSGHHHSARMFCDITMP
Rv2222c|M.tuberculosis_H37Rv FDGSSIEGFARVSESDTVAHPDPSTFQVLPWATSSGHHHSARMFCDITMP
MMAR_3294|M.marinum_M FDGSSIEGFARVSESDTVAHPDPSTFQVLPWVTDAGHHHSARIFCDITMP
MUL_1335|M.ulcerans_Agy99 FDGSSIEGFARVSESDTVAHPDPSTFQVLPWVTDAGHHHSARIFCDITMP
MAV_2244|M.avium_104 FDGSSIEGFSRVSESDTVANPDPSTFQVLPWASPNGHHHSARMFCDITMP
MLBr_01631|M.leprae_Br4923 FDGSSIEGFARVSESDTVAHPDPSTFQILPWATTAGHHHSARMFCDITMP
MAB_1920|M.abscessus_ATCC_1997 FDGSSIEGFARVSESDTVAHPDPSTFQVLPWTTSAGKHHSARMFCDIKLP
****:****:** *:* **.*******:*** *:*:***:****.:*
Mflv_2909|M.gilvum_PYR-GCK DGSPSWADSRHVLRRQLSKASDLGFSCYVHPEIEFFLLKPGDNDG--SPP
Mvan_3598|M.vanbaalenii_PYR-1 DGSPSWADSRHVLRRQLSKASDLGFSCYVHPEIEFFLLKPGPDDG--TQP
MSMEG_4294|M.smegmatis_MC2_155 DGSPSWADSRHVLRRQLAKASDLGFTCYVHPEIEFFLLKPGPNDG--TPP
TH_3396|M.thermoresistible__bu DGSPSWADPRHVLRRQLAKAGDQGFTCYVHPEIEFFLLKPGPHDG--SPP
Mb2246c|M.bovis_AF2122/97 DGSPSWADPRHVLRRQLTKAGELGFSCYVHPEIEFFLLKPGPEDG--SVP
Rv2222c|M.tuberculosis_H37Rv DGSPSWADPRHVLRRQLTKAGELGFSCYVHPEIEFFLLKPGPEDG--SVP
MMAR_3294|M.marinum_M DGSPSWADPRHVLRRQLTKANELGFSCYVHPEIEFFLLNPGPEDG--TEP
MUL_1335|M.ulcerans_Agy99 DGSPSWADPRHVLRRQLIKANELGFSCYVHPEIEFFLLNPGPEDG--TEP
MAV_2244|M.avium_104 DGSPSWADPRHVLRRQLQKANDLGFSCYVHPEIEFFLLKPGPDDG--TPP
MLBr_01631|M.leprae_Br4923 DGSSSWADPRHVLRRQLTKANDLGFSCYVHPEIEFFLLKAGPEDGSMPIP
MAB_1920|M.abscessus_ATCC_1997 DGSPSWSDPRHVLRRQLAKASDLGFSCYVHPEIEFFLLKNLPDDG--TAP
***.**:*.******** **.: **:************: .** . *
Mflv_2909|M.gilvum_PYR-GCK TPADNGGYFDQAVHDSAPNFRRHAIDALEQMGISVEFSHHEGAPGQQEID
Mvan_3598|M.vanbaalenii_PYR-1 IPADNGGYFDQAVHDAAPNFRRHAIDALEQMGISVEFSHHEGAPGQQEID
MSMEG_4294|M.smegmatis_MC2_155 EPADNGGYFDQAVHDAAPNFRRHAIEALEQMGISVEFSHHEGAPGQQEID
TH_3396|M.thermoresistible__bu VPADTSGYFDQSVHDTAPNFRRHAIEALEQMGISVEFSHHEGAPGQQEID
Mb2246c|M.bovis_AF2122/97 VPVDNAGYFDQAVHDSALNFRRHAIDALEFMGISVEFSHHEGAPGQQEID
Rv2222c|M.tuberculosis_H37Rv VPVDNAGYFDQAVHDSALNFRRHAIDALEFMGISVEFSHHEGAPGQQEID
MMAR_3294|M.marinum_M VPIDNAGYFDQAVHVSASKFRRHAIDALEFMGISVEFSHHEGAPGQQEID
MUL_1335|M.ulcerans_Agy99 VPIDNAGYFDQAVHVSASKFRRHVIDALEFMGISVEFSHHEGAPGQQEID
MAV_2244|M.avium_104 VPVDTAGYFDQAIHDSASNFRRHAIEALEFMGISVEFSHHEGAPGQQEID
MLBr_01631|M.leprae_Br4923 VPVDNAGYFDQAVHDSALNFRRHAIEALEFMGISVEFSHHEGAPGQQEID
MAB_1920|M.abscessus_ATCC_1997 IPADDAGYFDQAVHEAAPNFRRHAIDALESMGISVEFSHHEGAPGQQEID
* * .*****::* :* :****.*:*** ********************
Mflv_2909|M.gilvum_PYR-GCK LRYADALSMADNVMTFRYLVKEVALGDGVRASFMPKPFAEHPGSAMHTHM
Mvan_3598|M.vanbaalenii_PYR-1 LRYADALSMADNVMTFRYLVKEVALADGVRASFMPKPFAEHPGSAMHTHM
MSMEG_4294|M.smegmatis_MC2_155 LRYADALSMADNVMTFRYLVKEVALADGVRASFMPKPFAEHPGSAMHTHM
TH_3396|M.thermoresistible__bu LRYADALSMADNVMTFRYVVKQVAISDGVRASFMPKPFAEHPGSAMHTHM
Mb2246c|M.bovis_AF2122/97 LRFADALSMADNVMTFRYVIKEVALEEGARASFMPKPFGQHPGSAMHTHM
Rv2222c|M.tuberculosis_H37Rv LRFADALSMADNVMTFRYVIKEVALEEGARASFMPKPFGQHPGSAMHTHM
MMAR_3294|M.marinum_M LRFADALSMADNVMTFRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHM
MUL_1335|M.ulcerans_Agy99 LRFADALSMADNVMTFRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHM
MAV_2244|M.avium_104 LRFADALSMADNVMTFRYVIKEVAIENGARASFMPKPFGQHPGSAMHTHM
MLBr_01631|M.leprae_Br4923 LRFADALSMADNVMTFRYVIKEVAIENGARASFMPKPFAQHPGSAMHTHM
MAB_1920|M.abscessus_ATCC_1997 LRYADALSMADNVMTFRNVVKEVAIIDGVHATFMPKPFSDHPGSAMHTHM
**:************** ::*:**: :*.:*:******.:*.********
Mflv_2909|M.gilvum_PYR-GCK SLFEGDSNAFHSPDDPLQLSAVGKSFIAGILEHANEISAVTNQWVNSYKR
Mvan_3598|M.vanbaalenii_PYR-1 SLFEGDTNAFHSPDDPLQLSDVGKSFIAGILEHANEISAVTNQWVNSYKR
MSMEG_4294|M.smegmatis_MC2_155 SLFEGDTNAFHSPDDPLQLSDVAKSFIAGILEHANEISAVTNQWVNSYKR
TH_3396|M.thermoresistible__bu SLFEGDTNAFHSPDDPLQLSDVAKSFIAGILEHANEISAVTNQWVNSYKR
Mb2246c|M.bovis_AF2122/97 SLFEGDVNAFHSADDPLQLSEVGKSFIAGILEHACEISAVTNQWVNSYKR
Rv2222c|M.tuberculosis_H37Rv SLFEGDVNAFHSADDPLQLSEVGKSFIAGILEHACEISAVTNQWVNSYKR
MMAR_3294|M.marinum_M SLFEGDVNAFHSPDDPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKR
MUL_1335|M.ulcerans_Agy99 SLFEGDVNAFHSPDDPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKR
MAV_2244|M.avium_104 SLFEGDVNAFHSPDDPLQLSDVGKSFIAGILEHASEISAVTNQWVNSYKR
MLBr_01631|M.leprae_Br4923 SLFEGDVNAFHSDDDPLQLSDVGKSFIAGILEHASEISAVTNQWVNSYKR
MAB_1920|M.abscessus_ATCC_1997 SLFEGDTNAFHNPDDPLQLSDVGKSFIAGILEHANEISAVTNQWVNSYKR
****** ****. ******* *.*********** ***************
Mflv_2909|M.gilvum_PYR-GCK LVHGGEAPTAASWGAANRSALVRVPMYTPHKVSSRRVEVRSPDSACNPYL
Mvan_3598|M.vanbaalenii_PYR-1 LVHGGEAPTAASWGAANRSALVRVPMYTPHKVSSRRVEVRSPDSACNPYL
MSMEG_4294|M.smegmatis_MC2_155 LVHGGEAPTAASWGAANRSALVRVPMYTPHKVSSRRVEVRSPDSACNPYL
TH_3396|M.thermoresistible__bu LVHGGEAPTAASWGAANRSALVRVPMYTPHKASSRRVEVRSPDSACNPYL
Mb2246c|M.bovis_AF2122/97 LVQGGEAPTAASWGAANRSALVRVPMYTPHKTSSRRVEVRSPDSACNPYL
Rv2222c|M.tuberculosis_H37Rv LVQGGEAPTAASWGAANRSALVRVPMYTPHKTSSRRVEVRSPDSACNPYL
MMAR_3294|M.marinum_M LILGGEAPTAASWGAANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYL
MUL_1335|M.ulcerans_Agy99 LILGGEAPTAASWGAANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYL
MAV_2244|M.avium_104 LVGGGEAPTAASWGAANRSALVRVPMYTPHKTSSRRVEVRSPDSACNPYL
MLBr_01631|M.leprae_Br4923 LVVGGEAPTTASWGAANRSALVRVPMYTPHKTSSRRVEVRSPDSACNPYL
MAB_1920|M.abscessus_ATCC_1997 LVQGGEAPTAASWGAANRSALVRVPMYTPNKSSSRRIEVRSPDSACNPYL
*: ******:*******************:* ****:*************
Mflv_2909|M.gilvum_PYR-GCK TFAVLLAAGLRGIEKNYVLGPEAEDNVWSLTPEERRAMGYKELPGSLGAA
Mvan_3598|M.vanbaalenii_PYR-1 TFAVLLAAGLRGIEKNYVLGPQAEDNVWSLTPEERRAMGYKELPGSLGVA
MSMEG_4294|M.smegmatis_MC2_155 TFAVLLAAGLRGVEKGYVLGPQAEDNVWSLTQEERRAMGYRELPTSLGNA
TH_3396|M.thermoresistible__bu TFAVLLAAGLRGVEKGYVLGPEAEDNVWTLTPEERRAMGFKELPSSLGQA
Mb2246c|M.bovis_AF2122/97 TFAVLLAAGLRGVEKGYVLGPQAEDNVWDLTPEERRAMGYRELPSSLDSA
Rv2222c|M.tuberculosis_H37Rv TFAVLLAAGLRGVEKGYVLGPQAEDNVWDLTPEERRAMGYRELPSSLDSA
MMAR_3294|M.marinum_M AFAVLLAAGLRGVEKGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSA
MUL_1335|M.ulcerans_Agy99 AFAVLLAAGLRGVEKGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSA
MAV_2244|M.avium_104 TFAVLLAAGLRGVEKGYVLGPQAEDNVWDLTPEERIAMGYRELPTSLDSA
MLBr_01631|M.leprae_Br4923 AFAVLLAAGLRGVEKGYVLGPQAEDNVWDLTPEERLAMGYRELPTSLDSA
MAB_1920|M.abscessus_ATCC_1997 TYAVLLAAGLRGVEQGYTLGPEAEDDVWGLTRAERQAMGYKELPSTLENA
::**********:*:.*.***:***:** ** ** :**::*** :* *
Mflv_2909|M.gilvum_PYR-GCK LADMENSELVAEALGEHVFDFFLRNKRREWENYRSHVTPFELKNYLSL
Mvan_3598|M.vanbaalenii_PYR-1 LTEMENSELVAEALGEHVFDFFLRNKRSEWENYRSHVTPFELKNYLSL
MSMEG_4294|M.smegmatis_MC2_155 LESMENSELVAEALGEHVFDYFLRNKRSEWENYRSHVTPYELKNYLSL
TH_3396|M.thermoresistible__bu LAEMERSELVAEALGEHVFDFFLRNKRAEWENYRSHVTPYELKNYLSL
Mb2246c|M.bovis_AF2122/97 LRAMEASELVAEALGEHVFDFFLRNKRTEWANYRSHVTPYELRTYLSL
Rv2222c|M.tuberculosis_H37Rv LRAMEASELVAEALGEHVFDFFLRNKRTEWANYRSHVTPYELRTYLSL
MMAR_3294|M.marinum_M LRAMESSELVAEALGEHVFDFFLRNKRREWATYRSHVTPYELSTYLSL
MUL_1335|M.ulcerans_Agy99 LRAMESSELVAEALGEHVFDFFLRNKRREWATYRSHVTPYELSTYLSL
MAV_2244|M.avium_104 LRAMESSELVAETLGEHVFDFFLRNKRTEWATYRSHVTPYELRTYLSL
MLBr_01631|M.leprae_Br4923 LGAMEVSEFVAEALGEHVFDFFLRNKRAEWANYRSHVSLYELKTYLSL
MAB_1920|M.abscessus_ATCC_1997 LIAMEGSELVAEALGEHVFDFFLRNKRAEWARYRSNVTPFELRAYLGL
* ** **:***:*******:****** ** ***:*: :** **.*